Matthieu Darcy
Solving Functional PDEs with Gaussian Processes and Applications to Functional Renormalization Group Equations
Dec 24, 2025Abstract:We present an operator learning framework for solving non-perturbative functional renormalization group equations, which are integro-differential equations defined on functionals. Our proposed approach uses Gaussian process operator learning to construct a flexible functional representation formulated directly on function space, making it independent of a particular equation or discretization. Our method is flexible, and can apply to a broad range of functional differential equations while still allowing for the incorporation of physical priors in either the prior mean or the kernel design. We demonstrate the performance of our method on several relevant equations, such as the Wetterich and Wilson--Polchinski equations, showing that it achieves equal or better performance than existing approximations such as the local-potential approximation, while being significantly more flexible. In particular, our method can handle non-constant fields, making it promising for the study of more complex field configurations, such as instantons.
Solving Roughly Forced Nonlinear PDEs via Misspecified Kernel Methods and Neural Networks
Jan 29, 2025Abstract:We consider the use of Gaussian Processes (GPs) or Neural Networks (NNs) to numerically approximate the solutions to nonlinear partial differential equations (PDEs) with rough forcing or source terms, which commonly arise as pathwise solutions to stochastic PDEs. Kernel methods have recently been generalized to solve nonlinear PDEs by approximating their solutions as the maximum a posteriori estimator of GPs that are conditioned to satisfy the PDE at a finite set of collocation points. The convergence and error guarantees of these methods, however, rely on the PDE being defined in a classical sense and its solution possessing sufficient regularity to belong to the associated reproducing kernel Hilbert space. We propose a generalization of these methods to handle roughly forced nonlinear PDEs while preserving convergence guarantees with an oversmoothing GP kernel that is misspecified relative to the true solution's regularity. This is achieved by conditioning a regular GP to satisfy the PDE with a modified source term in a weak sense (when integrated against a finite number of test functions). This is equivalent to replacing the empirical $L^2$-loss on the PDE constraint by an empirical negative-Sobolev norm. We further show that this loss function can be used to extend physics-informed neural networks (PINNs) to stochastic equations, thereby resulting in a new NN-based variant termed Negative Sobolev Norm-PINN (NeS-PINN).
Kernel Methods are Competitive for Operator Learning
Apr 26, 2023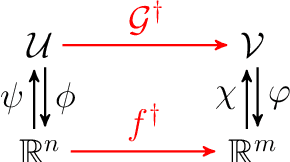
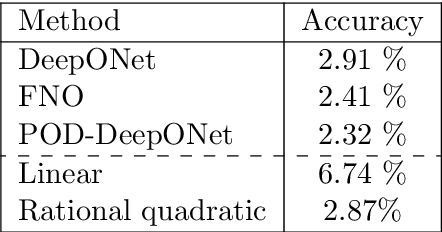
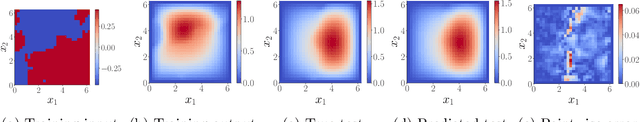
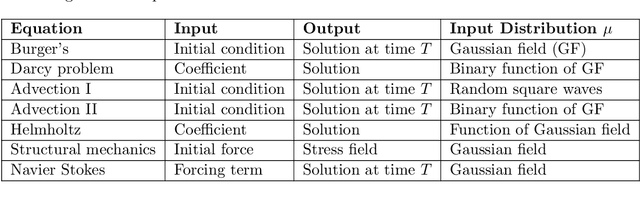
Abstract:We present a general kernel-based framework for learning operators between Banach spaces along with a priori error analysis and comprehensive numerical comparisons with popular neural net (NN) approaches such as Deep Operator Net (DeepONet) [Lu et al.] and Fourier Neural Operator (FNO) [Li et al.]. We consider the setting where the input/output spaces of target operator $\mathcal{G}^\dagger\,:\, \mathcal{U}\to \mathcal{V}$ are reproducing kernel Hilbert spaces (RKHS), the data comes in the form of partial observations $\phi(u_i), \varphi(v_i)$ of input/output functions $v_i=\mathcal{G}^\dagger(u_i)$ ($i=1,\ldots,N$), and the measurement operators $\phi\,:\, \mathcal{U}\to \mathbb{R}^n$ and $\varphi\,:\, \mathcal{V} \to \mathbb{R}^m$ are linear. Writing $\psi\,:\, \mathbb{R}^n \to \mathcal{U}$ and $\chi\,:\, \mathbb{R}^m \to \mathcal{V}$ for the optimal recovery maps associated with $\phi$ and $\varphi$, we approximate $\mathcal{G}^\dagger$ with $\bar{\mathcal{G}}=\chi \circ \bar{f} \circ \phi$ where $\bar{f}$ is an optimal recovery approximation of $f^\dagger:=\varphi \circ \mathcal{G}^\dagger \circ \psi\,:\,\mathbb{R}^n \to \mathbb{R}^m$. We show that, even when using vanilla kernels (e.g., linear or Mat\'{e}rn), our approach is competitive in terms of cost-accuracy trade-off and either matches or beats the performance of NN methods on a majority of benchmarks. Additionally, our framework offers several advantages inherited from kernel methods: simplicity, interpretability, convergence guarantees, a priori error estimates, and Bayesian uncertainty quantification. As such, it can serve as a natural benchmark for operator learning.
One-Shot Learning of Stochastic Differential Equations with Computational Graph Completion
Sep 24, 2022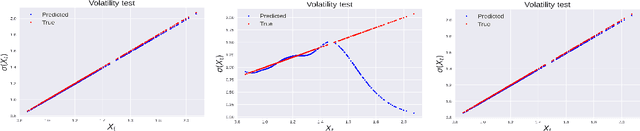
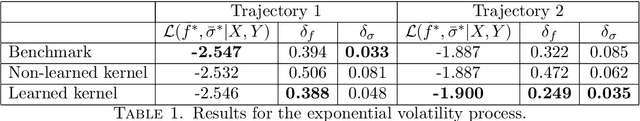
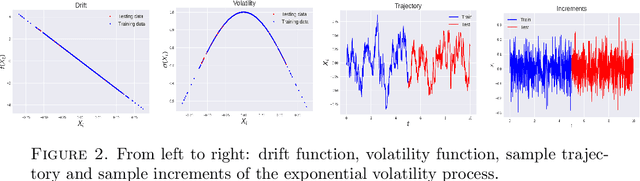
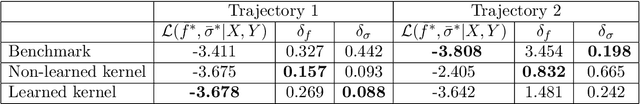
Abstract:We consider the problem of learning Stochastic Differential Equations of the form $dX_t = f(X_t)dt+\sigma(X_t)dW_t $ from one sample trajectory. This problem is more challenging than learning deterministic dynamical systems because one sample trajectory only provides indirect information on the unknown functions $f$, $\sigma$, and stochastic process $dW_t$ representing the drift, the diffusion, and the stochastic forcing terms, respectively. We propose a simple kernel-based solution to this problem that can be decomposed as follows: (1) Represent the time-increment map $X_t \rightarrow X_{t+dt}$ as a Computational Graph in which $f$, $\sigma$ and $dW_t$ appear as unknown functions and random variables. (2) Complete the graph (approximate unknown functions and random variables) via Maximum a Posteriori Estimation (given the data) with Gaussian Process (GP) priors on the unknown functions. (3) Learn the covariance functions (kernels) of the GP priors from data with randomized cross-validation. Numerical experiments illustrate the efficacy, robustness, and scope of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge