Daniel Ratner
Efficient Nudged Elastic Band Method using Neural Network Bayesian Algorithm Execution
Dec 17, 2025Abstract:The discovery of a minimum energy pathway (MEP) between metastable states is crucial for scientific tasks including catalyst and biomolecular design. However, the standard nudged elastic band (NEB) algorithm requires hundreds to tens of thousands of compute-intensive simulations, making applications to complex systems prohibitively expensive. We introduce Neural Network Bayesian Algorithm Execution (NN-BAX), a framework that jointly learns the energy landscape and the MEP. NN-BAX sequentially fine-tunes a foundation model by actively selecting samples targeted at improving the MEP. Tested on Lennard-Jones and Embedded Atom Method systems, our approach achieves a one to two order of magnitude reduction in energy and force evaluations with negligible loss in MEP accuracy and demonstrates scalability to >100-dimensional systems. This work is therefore a promising step towards removing the computational barrier for MEP discovery in scientifically relevant systems, suggesting that weeks-long calculations may be achieved in hours or days with minimal loss in accuracy.
eLog analysis for accelerators: status and future outlook
Jun 15, 2025
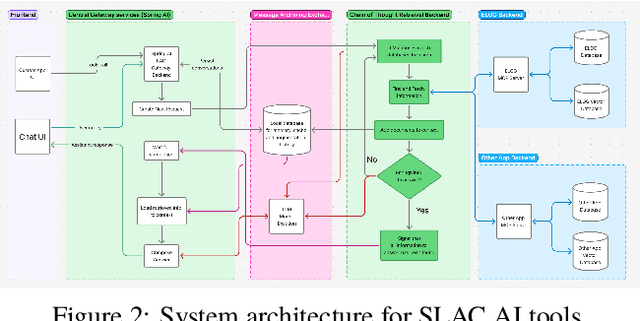
Abstract:This work demonstrates electronic logbook (eLog) systems leveraging modern AI-driven information retrieval capabilities at the accelerator facilities of Fermilab, Jefferson Lab, Lawrence Berkeley National Laboratory (LBNL), SLAC National Accelerator Laboratory. We evaluate contemporary tools and methodologies for information retrieval with Retrieval Augmented Generation (RAGs), focusing on operational insights and integration with existing accelerator control systems. The study addresses challenges and proposes solutions for state-of-the-art eLog analysis through practical implementations, demonstrating applications and limitations. We present a framework for enhancing accelerator facility operations through improved information accessibility and knowledge management, which could potentially lead to more efficient operations.
A Start To End Machine Learning Approach To Maximize Scientific Throughput From The LCLS-II-HE
May 29, 2025Abstract:With the increasing brightness of Light sources, including the Diffraction-Limited brightness upgrade of APS and the high-repetition-rate upgrade of LCLS, the proposed experiments therein are becoming increasingly complex. For instance, experiments at LCLS-II-HE will require the X-ray beam to be within a fraction of a micron in diameter, with pointing stability of a few nanoradians, at the end of a kilometer-long electron accelerator, a hundred-meter-long undulator section, and tens of meters long X-ray optics. This enhancement of brightness will increase the data production rate to rival the largest data generators in the world. Without real-time active feedback control and an optimized pipeline to transform measurements to scientific information and insights, researchers will drown in a deluge of mostly useless data, and fail to extract the highly sophisticated insights that the recent brightness upgrades promise. In this article, we outline the strategy we are developing at SLAC to implement Machine Learning driven optimization, automation and real-time knowledge extraction from the electron-injector at the start of the electron accelerator, to the multidimensional X-ray optical systems, and till the experimental endstations and the high readout rate, multi-megapixel detectors at LCLS to deliver the design performance to the users. This is illustrated via examples from Accelerator, Optics and End User applications.
Resilient VAE: Unsupervised Anomaly Detection at the SLAC Linac Coherent Light Source
Sep 05, 2023Abstract:Significant advances in utilizing deep learning for anomaly detection have been made in recent years. However, these methods largely assume the existence of a normal training set (i.e., uncontaminated by anomalies) or even a completely labeled training set. In many complex engineering systems, such as particle accelerators, labels are sparse and expensive; in order to perform anomaly detection in these cases, we must drop these assumptions and utilize a completely unsupervised method. This paper introduces the Resilient Variational Autoencoder (ResVAE), a deep generative model specifically designed for anomaly detection. ResVAE exhibits resilience to anomalies present in the training data and provides feature-level anomaly attribution. During the training process, ResVAE learns the anomaly probability for each sample as well as each individual feature, utilizing these probabilities to effectively disregard anomalous examples in the training data. We apply our proposed method to detect anomalies in the accelerator status at the SLAC Linac Coherent Light Source (LCLS). By utilizing shot-to-shot data from the beam position monitoring system, we demonstrate the exceptional capability of ResVAE in identifying various types of anomalies that are visible in the accelerator.
Capturing dynamical correlations using implicit neural representations
Apr 08, 2023Abstract:The observation and description of collective excitations in solids is a fundamental issue when seeking to understand the physics of a many-body system. Analysis of these excitations is usually carried out by measuring the dynamical structure factor, S(Q, $\omega$), with inelastic neutron or x-ray scattering techniques and comparing this against a calculated dynamical model. Here, we develop an artificial intelligence framework which combines a neural network trained to mimic simulated data from a model Hamiltonian with automatic differentiation to recover unknown parameters from experimental data. We benchmark this approach on a Linear Spin Wave Theory (LSWT) simulator and advanced inelastic neutron scattering data from the square-lattice spin-1 antiferromagnet La$_2$NiO$_4$. We find that the model predicts the unknown parameters with excellent agreement relative to analytical fitting. In doing so, we illustrate the ability to build and train a differentiable model only once, which then can be applied in real-time to multi-dimensional scattering data, without the need for human-guided peak finding and fitting algorithms. This prototypical approach promises a new technology for this field to automatically detect and refine more advanced models for ordered quantum systems.
Coincident Learning for Unsupervised Anomaly Detection
Jan 26, 2023



Abstract:Anomaly detection is an important task for complex systems (e.g., industrial facilities, manufacturing, large-scale science experiments), where failures in a sub-system can lead to low yield, faulty products, or even damage to components. While complex systems often have a wealth of data, labeled anomalies are typically rare (or even nonexistent) and expensive to acquire. In this paper, we introduce a new method, called CoAD, for training anomaly detection models on unlabeled data, based on the expectation that anomalous behavior in one sub-system will produce coincident anomalies in downstream sub-systems and products. Given data split into two streams $s$ and $q$ (i.e., subsystem diagnostics and final product quality), we define an unsupervised metric, $\hat{F}_\beta$, out of analogy to the supervised classification $F_\beta$ statistic, which quantifies the performance of the independent anomaly detection algorithms on s and q based on their coincidence rate. We demonstrate our method in four cases: a synthetic time-series data set, a synthetic imaging data set generated from MNIST, a metal milling data set, and a data set taken from a particle accelerator.
Implicit Neural Representation as a Differentiable Surrogate for Photon Propagation in a Monolithic Neutrino Detector
Nov 02, 2022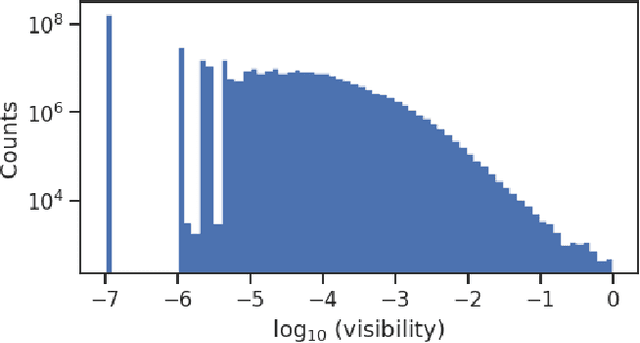
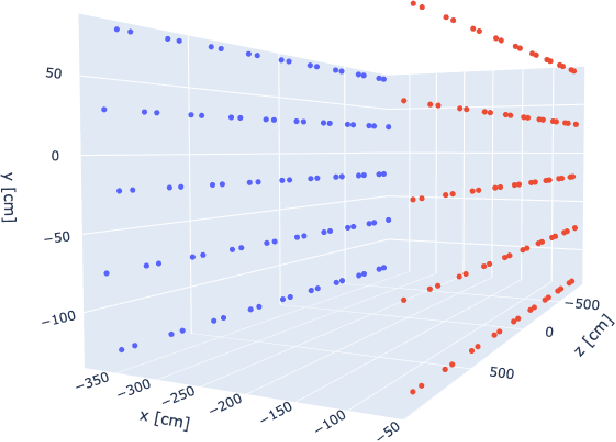
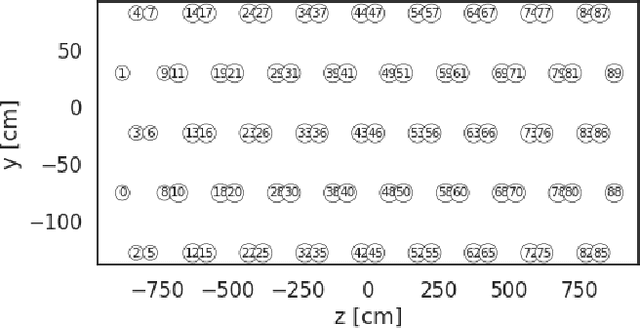
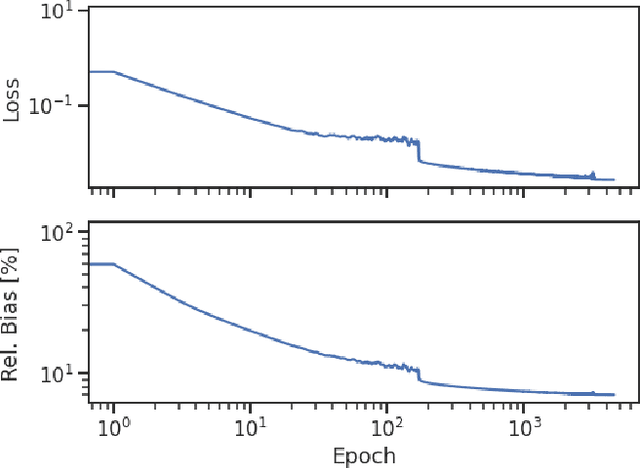
Abstract:Optical photons are used as signal in a wide variety of particle detectors. Modern neutrino experiments employ hundreds to tens of thousands of photon detectors to observe signal from millions to billions of scintillation photons produced from energy deposition of charged particles. These neutrino detectors are typically large, containing kilotons of target volume, with different optical properties. Modeling individual photon propagation in form of look-up table requires huge computational resources. As the size of a table increases with detector volume for a fixed resolution, this method scales poorly for future larger detectors. Alternative approaches such as fitting a polynomial to the model could address the memory issue, but results in poorer performance. Both look-up table and fitting approaches are prone to discrepancies between the detector simulation and the data collected. We propose a new approach using SIREN, an implicit neural representation with periodic activation functions, to model the look-up table as a 3D scene and reproduces the acceptance map with high accuracy. The number of parameters in our SIREN model is orders of magnitude smaller than the number of voxels in the look-up table. As it models an underlying functional shape, SIREN is scalable to a larger detector. Furthermore, SIREN can successfully learn the spatial gradients of the photon library, providing additional information for downstream applications. Finally, as SIREN is a neural network representation, it is differentiable with respect to its parameters, and therefore tunable via gradient descent. We demonstrate the potential of optimizing SIREN directly on real data, which mitigates the concern of data vs. simulation discrepancies. We further present an application for data reconstruction where SIREN is used to form a likelihood function for photon statistics.
Heterogeneous reconstruction of deformable atomic models in Cryo-EM
Sep 29, 2022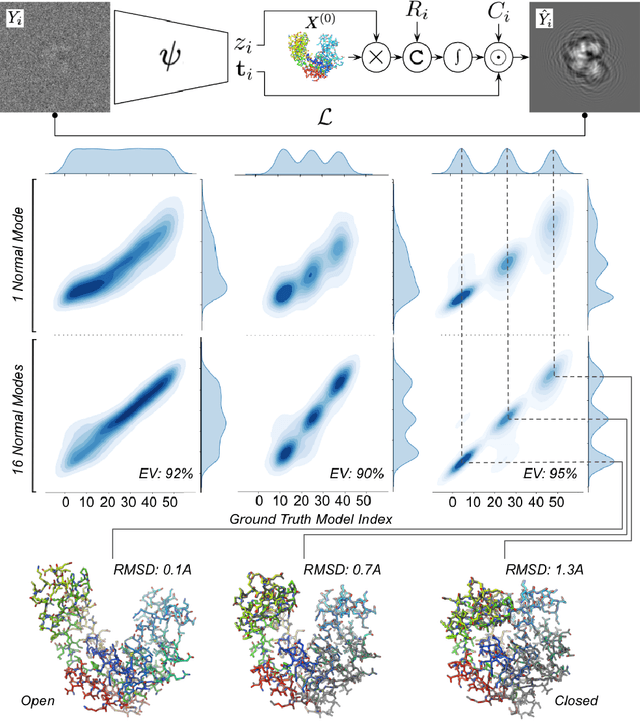
Abstract:Cryogenic electron microscopy (cryo-EM) provides a unique opportunity to study the structural heterogeneity of biomolecules. Being able to explain this heterogeneity with atomic models would help our understanding of their functional mechanisms but the size and ruggedness of the structural space (the space of atomic 3D cartesian coordinates) presents an immense challenge. Here, we describe a heterogeneous reconstruction method based on an atomistic representation whose deformation is reduced to a handful of collective motions through normal mode analysis. Our implementation uses an autoencoder. The encoder jointly estimates the amplitude of motion along the normal modes and the 2D shift between the center of the image and the center of the molecule . The physics-based decoder aggregates a representation of the heterogeneity readily interpretable at the atomic level. We illustrate our method on 3 synthetic datasets corresponding to different distributions along a simulated trajectory of adenylate kinase transitioning from its open to its closed structures. We show for each distribution that our approach is able to recapitulate the intermediate atomic models with atomic-level accuracy.
Bayesian Algorithm Execution for Tuning Particle Accelerator Emittance with Partial Measurements
Sep 10, 2022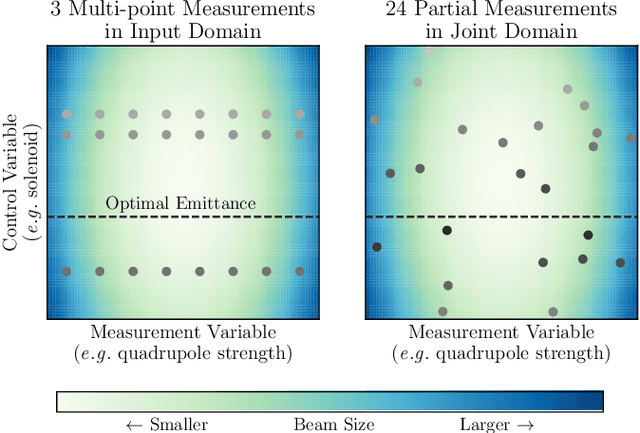
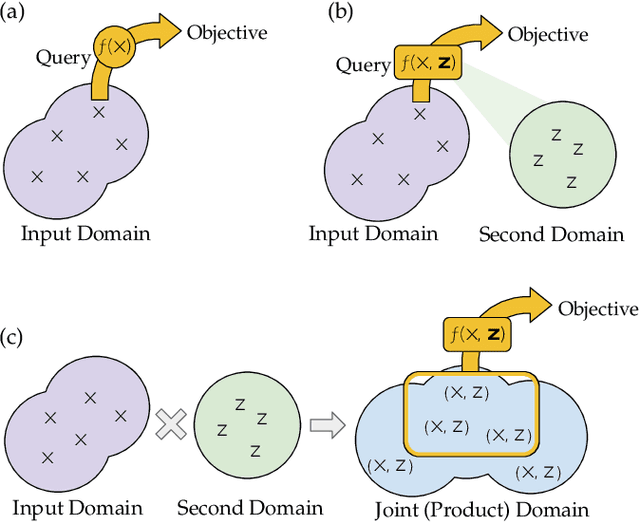
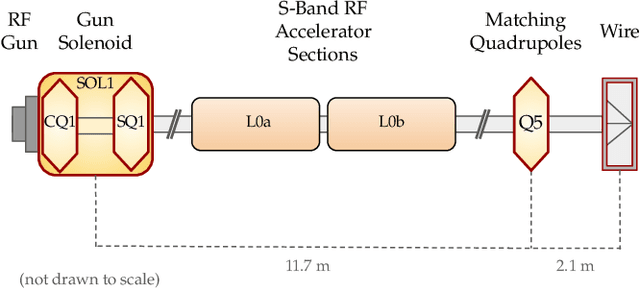
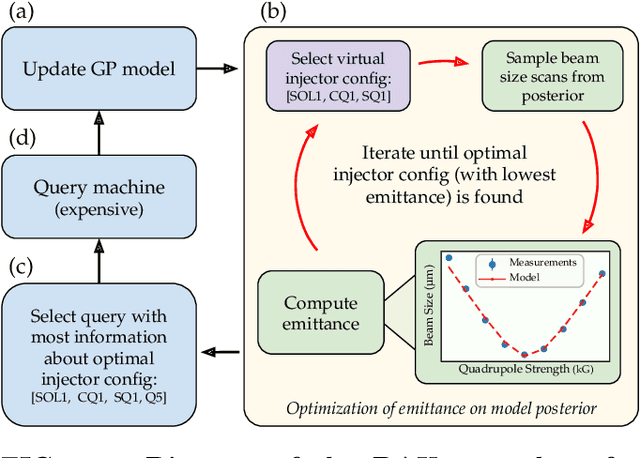
Abstract:Traditional black-box optimization methods are inefficient when dealing with multi-point measurement, i.e. when each query in the control domain requires a set of measurements in a secondary domain to calculate the objective. In particle accelerators, emittance tuning from quadrupole scans is an example of optimization with multi-point measurements. Although the emittance is a critical parameter for the performance of high-brightness machines, including X-ray lasers and linear colliders, comprehensive optimization is often limited by the time required for tuning. Here, we extend the recently-proposed Bayesian Algorithm Execution (BAX) to the task of optimization with multi-point measurements. BAX achieves sample-efficiency by selecting and modeling individual points in the joint control-measurement domain. We apply BAX to emittance minimization at the Linac Coherent Light Source (LCLS) and the Facility for Advanced Accelerator Experimental Tests II (FACET-II) particle accelerators. In an LCLS simulation environment, we show that BAX delivers a 20x increase in efficiency while also being more robust to noise compared to traditional optimization methods. Additionally, we ran BAX live at both LCLS and FACET-II, matching the hand-tuned emittance at FACET-II and achieving an optimal emittance that was 24% lower than that obtained by hand-tuning at LCLS. We anticipate that our approach can readily be adapted to other types of optimization problems involving multi-point measurements commonly found in scientific instruments.
CryoAI: Amortized Inference of Poses for Ab Initio Reconstruction of 3D Molecular Volumes from Real Cryo-EM Images
Mar 16, 2022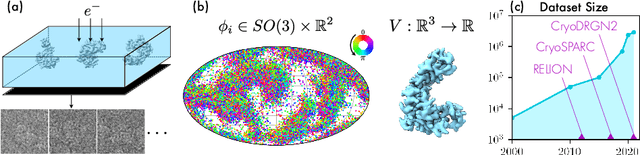
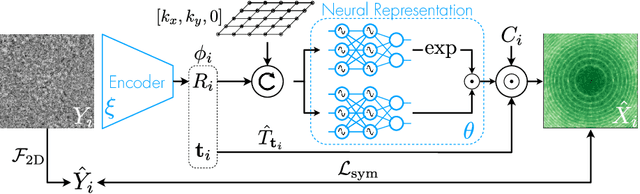
Abstract:Cryo-electron microscopy (cryo-EM) has become a tool of fundamental importance in structural biology, helping us understand the basic building blocks of life. The algorithmic challenge of cryo-EM is to jointly estimate the unknown 3D poses and the 3D electron scattering potential of a biomolecule from millions of extremely noisy 2D images. Existing reconstruction algorithms, however, cannot easily keep pace with the rapidly growing size of cryo-EM datasets due to their high computational and memory cost. We introduce cryoAI, an ab initio reconstruction algorithm for homogeneous conformations that uses direct gradient-based optimization of particle poses and the electron scattering potential from single-particle cryo-EM data. CryoAI combines a learned encoder that predicts the poses of each particle image with a physics-based decoder to aggregate each particle image into an implicit representation of the scattering potential volume. This volume is stored in the Fourier domain for computational efficiency and leverages a modern coordinate network architecture for memory efficiency. Combined with a symmetrized loss function, this framework achieves results of a quality on par with state-of-the-art cryo-EM solvers for both simulated and experimental data, one order of magnitude faster for large datasets and with significantly lower memory requirements than existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge