Geoffrey Woollard
CryoBench: Diverse and challenging datasets for the heterogeneity problem in cryo-EM
Aug 10, 2024



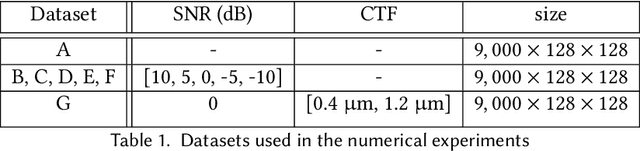
Abstract:Cryo-electron microscopy (cryo-EM) is a powerful technique for determining high-resolution 3D biomolecular structures from imaging data. As this technique can capture dynamic biomolecular complexes, 3D reconstruction methods are increasingly being developed to resolve this intrinsic structural heterogeneity. However, the absence of standardized benchmarks with ground truth structures and validation metrics limits the advancement of the field. Here, we propose CryoBench, a suite of datasets, metrics, and performance benchmarks for heterogeneous reconstruction in cryo-EM. We propose five datasets representing different sources of heterogeneity and degrees of difficulty. These include conformational heterogeneity generated from simple motions and random configurations of antibody complexes and from tens of thousands of structures sampled from a molecular dynamics simulation. We also design datasets containing compositional heterogeneity from mixtures of ribosome assembly states and 100 common complexes present in cells. We then perform a comprehensive analysis of state-of-the-art heterogeneous reconstruction tools including neural and non-neural methods and their sensitivity to noise, and propose new metrics for quantitative comparison of methods. We hope that this benchmark will be a foundational resource for analyzing existing methods and new algorithmic development in both the cryo-EM and machine learning communities.
End-to-End Simultaneous Learning of Single-particle Orientation and 3D Map Reconstruction from Cryo-electron Microscopy Data
Jul 07, 2021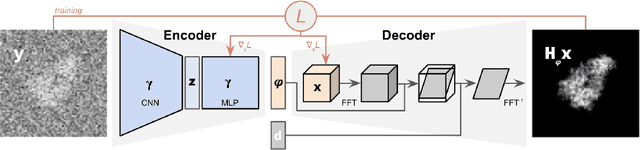
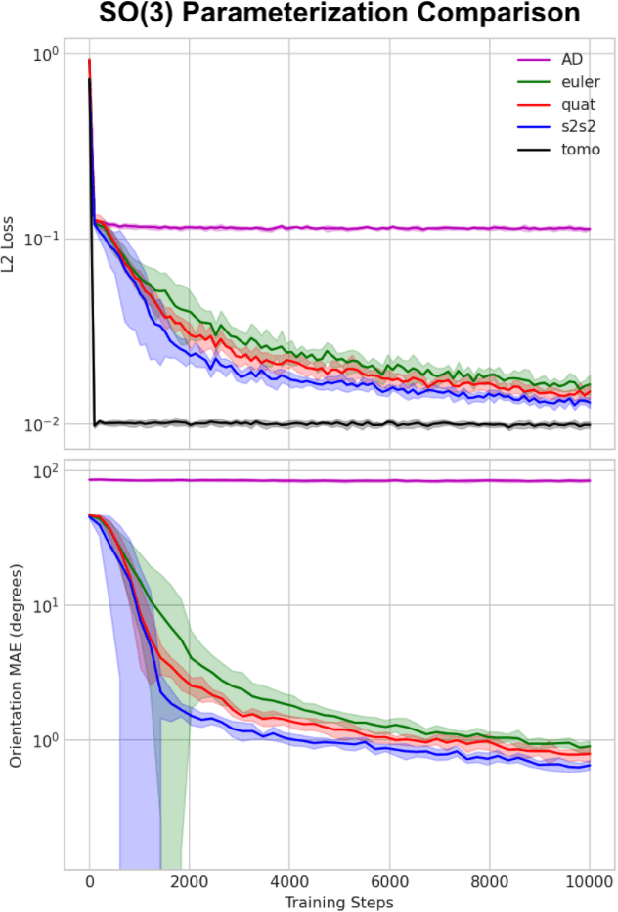
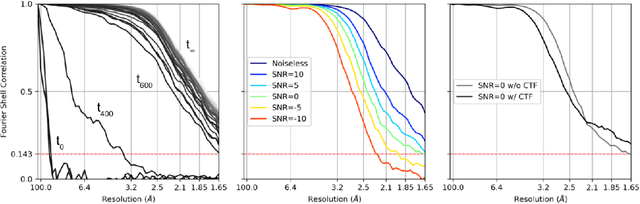
Abstract:Cryogenic electron microscopy (cryo-EM) provides images from different copies of the same biomolecule in arbitrary orientations. Here, we present an end-to-end unsupervised approach that learns individual particle orientations from cryo-EM data while reconstructing the average 3D map of the biomolecule, starting from a random initialization. The approach relies on an auto-encoder architecture where the latent space is explicitly interpreted as orientations used by the decoder to form an image according to the linear projection model. We evaluate our method on simulated data and show that it is able to reconstruct 3D particle maps from noisy- and CTF-corrupted 2D projection images of unknown particle orientations.
Transfer Learning for Risk Classification of Social Media Posts: Model Evaluation Study
Jul 10, 2019



Abstract:Mental illness affects a significant portion of the worldwide population. Online mental health forums can provide a supportive environment for those afflicted and also generate a large amount of data which can be mined to predict mental health states using machine learning methods. We benchmark multiple methods of text feature representation for social media posts and compare their downstream use with automated machine learning (AutoML) tools to triage content for moderator attention. We used 1588 labeled posts from the CLPsych 2017 shared task collected from the Reachout.com forum (Milne et al., 2019). Posts were represented using lexicon based tools including VADER, Empath, LIWC and also used pre-trained artificial neural network models including DeepMoji, Universal Sentence Encoder, and GPT-1. We used TPOT and auto-sklearn as AutoML tools to generate classifiers to triage the posts. The top-performing system used features derived from the GPT-1 model, which was finetuned on over 150,000 unlabeled posts from Reachout.com. Our top system had a macro averaged F1 score of 0.572, providing a new state-of-the-art result on the CLPsych 2017 task. This was achieved without additional information from meta-data or preceding posts. Error analyses revealed that this top system often misses expressions of hopelessness. We additionally present visualizations that aid understanding of the learned classifiers. We show that transfer learning is an effective strategy for predicting risk with relatively little labeled data. We note that finetuning of pretrained language models provides further gains when large amounts of unlabeled text is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge