End-to-End Simultaneous Learning of Single-particle Orientation and 3D Map Reconstruction from Cryo-electron Microscopy Data
Paper and Code
Jul 07, 2021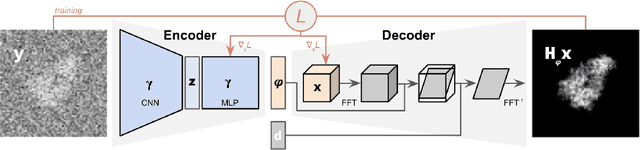
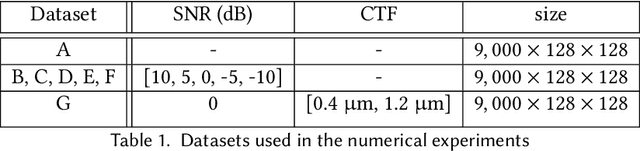
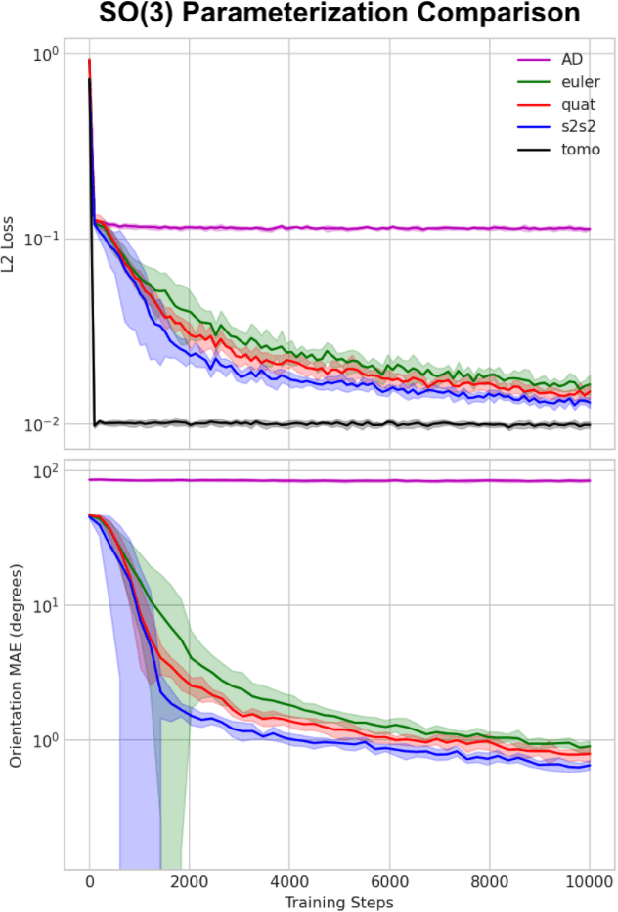
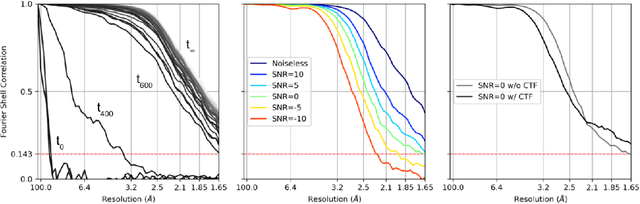
Cryogenic electron microscopy (cryo-EM) provides images from different copies of the same biomolecule in arbitrary orientations. Here, we present an end-to-end unsupervised approach that learns individual particle orientations from cryo-EM data while reconstructing the average 3D map of the biomolecule, starting from a random initialization. The approach relies on an auto-encoder architecture where the latent space is explicitly interpreted as orientations used by the decoder to form an image according to the linear projection model. We evaluate our method on simulated data and show that it is able to reconstruct 3D particle maps from noisy- and CTF-corrupted 2D projection images of unknown particle orientations.
* 13 pages, 4 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge