Minkyu Jeon
CryoBench: Diverse and challenging datasets for the heterogeneity problem in cryo-EM
Aug 10, 2024



Abstract:Cryo-electron microscopy (cryo-EM) is a powerful technique for determining high-resolution 3D biomolecular structures from imaging data. As this technique can capture dynamic biomolecular complexes, 3D reconstruction methods are increasingly being developed to resolve this intrinsic structural heterogeneity. However, the absence of standardized benchmarks with ground truth structures and validation metrics limits the advancement of the field. Here, we propose CryoBench, a suite of datasets, metrics, and performance benchmarks for heterogeneous reconstruction in cryo-EM. We propose five datasets representing different sources of heterogeneity and degrees of difficulty. These include conformational heterogeneity generated from simple motions and random configurations of antibody complexes and from tens of thousands of structures sampled from a molecular dynamics simulation. We also design datasets containing compositional heterogeneity from mixtures of ribosome assembly states and 100 common complexes present in cells. We then perform a comprehensive analysis of state-of-the-art heterogeneous reconstruction tools including neural and non-neural methods and their sensitivity to noise, and propose new metrics for quantitative comparison of methods. We hope that this benchmark will be a foundational resource for analyzing existing methods and new algorithmic development in both the cryo-EM and machine learning communities.
k-SALSA: k-anonymous synthetic averaging of retinal images via local style alignment
Mar 20, 2023Abstract:The application of modern machine learning to retinal image analyses offers valuable insights into a broad range of human health conditions beyond ophthalmic diseases. Additionally, data sharing is key to fully realizing the potential of machine learning models by providing a rich and diverse collection of training data. However, the personally-identifying nature of retinal images, encompassing the unique vascular structure of each individual, often prevents this data from being shared openly. While prior works have explored image de-identification strategies based on synthetic averaging of images in other domains (e.g. facial images), existing techniques face difficulty in preserving both privacy and clinical utility in retinal images, as we demonstrate in our work. We therefore introduce k-SALSA, a generative adversarial network (GAN)-based framework for synthesizing retinal fundus images that summarize a given private dataset while satisfying the privacy notion of k-anonymity. k-SALSA brings together state-of-the-art techniques for training and inverting GANs to achieve practical performance on retinal images. Furthermore, k-SALSA leverages a new technique, called local style alignment, to generate a synthetic average that maximizes the retention of fine-grain visual patterns in the source images, thus improving the clinical utility of the generated images. On two benchmark datasets of diabetic retinopathy (EyePACS and APTOS), we demonstrate our improvement upon existing methods with respect to image fidelity, classification performance, and mitigation of membership inference attacks. Our work represents a step toward broader sharing of retinal images for scientific collaboration. Code is available at https://github.com/hcholab/k-salsa.
SageMix: Saliency-Guided Mixup for Point Clouds
Oct 13, 2022
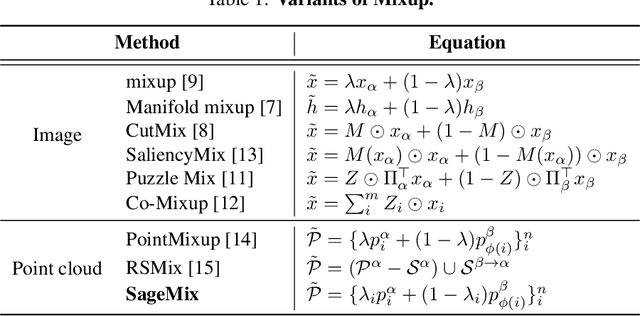
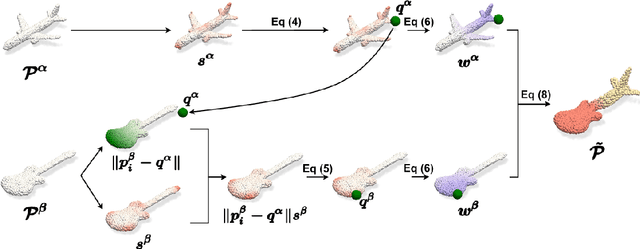
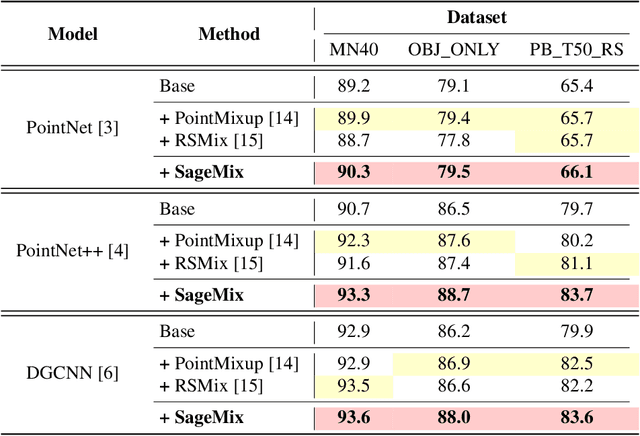
Abstract:Data augmentation is key to improving the generalization ability of deep learning models. Mixup is a simple and widely-used data augmentation technique that has proven effective in alleviating the problems of overfitting and data scarcity. Also, recent studies of saliency-aware Mixup in the image domain show that preserving discriminative parts is beneficial to improving the generalization performance. However, these Mixup-based data augmentations are underexplored in 3D vision, especially in point clouds. In this paper, we propose SageMix, a saliency-guided Mixup for point clouds to preserve salient local structures. Specifically, we extract salient regions from two point clouds and smoothly combine them into one continuous shape. With a simple sequential sampling by re-weighted saliency scores, SageMix preserves the local structure of salient regions. Extensive experiments demonstrate that the proposed method consistently outperforms existing Mixup methods in various benchmark point cloud datasets. With PointNet++, our method achieves an accuracy gain of 2.6% and 4.0% over standard training in 3D Warehouse dataset (MN40) and ScanObjectNN, respectively. In addition to generalization performance, SageMix improves robustness and uncertainty calibration. Moreover, when adopting our method to various tasks including part segmentation and standard 2D image classification, our method achieves competitive performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge