Youssef Nashed
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024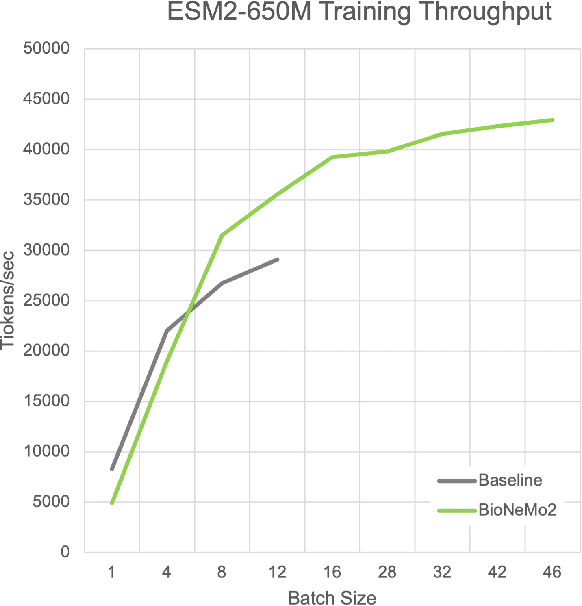
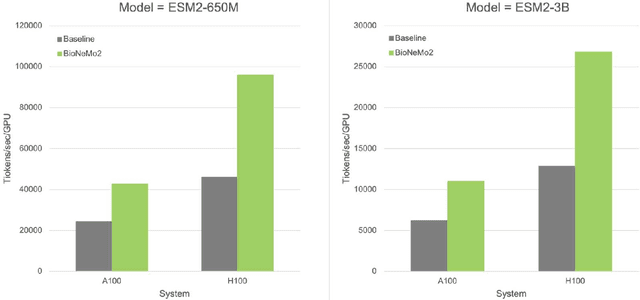
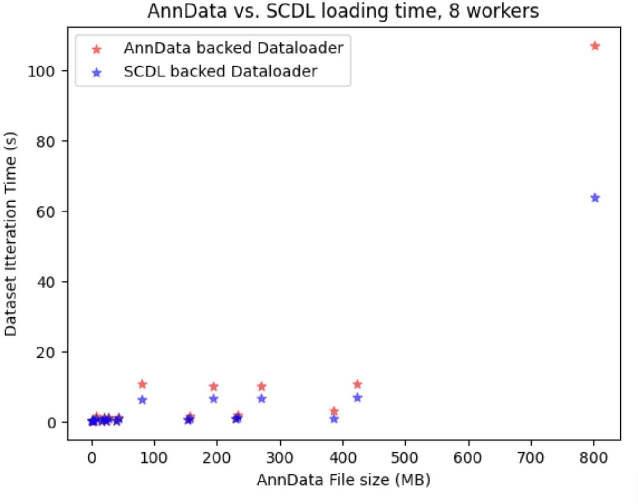
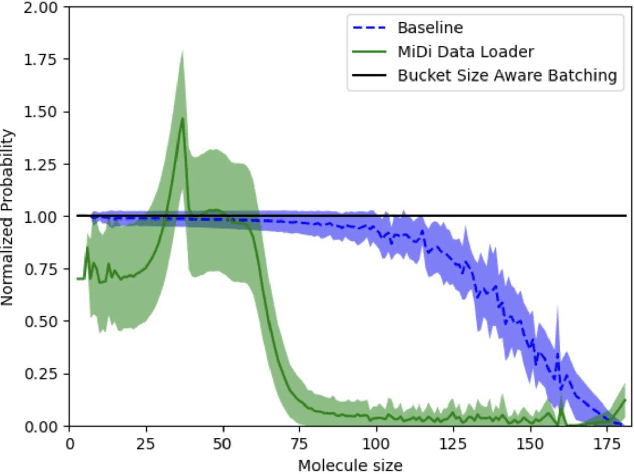
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
Capturing dynamical correlations using implicit neural representations
Apr 08, 2023Abstract:The observation and description of collective excitations in solids is a fundamental issue when seeking to understand the physics of a many-body system. Analysis of these excitations is usually carried out by measuring the dynamical structure factor, S(Q, $\omega$), with inelastic neutron or x-ray scattering techniques and comparing this against a calculated dynamical model. Here, we develop an artificial intelligence framework which combines a neural network trained to mimic simulated data from a model Hamiltonian with automatic differentiation to recover unknown parameters from experimental data. We benchmark this approach on a Linear Spin Wave Theory (LSWT) simulator and advanced inelastic neutron scattering data from the square-lattice spin-1 antiferromagnet La$_2$NiO$_4$. We find that the model predicts the unknown parameters with excellent agreement relative to analytical fitting. In doing so, we illustrate the ability to build and train a differentiable model only once, which then can be applied in real-time to multi-dimensional scattering data, without the need for human-guided peak finding and fitting algorithms. This prototypical approach promises a new technology for this field to automatically detect and refine more advanced models for ordered quantum systems.
Implicit Neural Representation as a Differentiable Surrogate for Photon Propagation in a Monolithic Neutrino Detector
Nov 02, 2022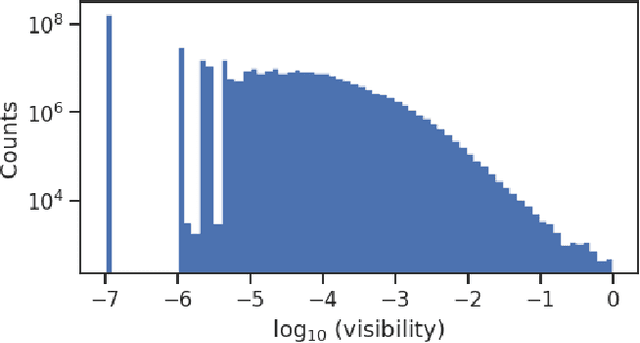
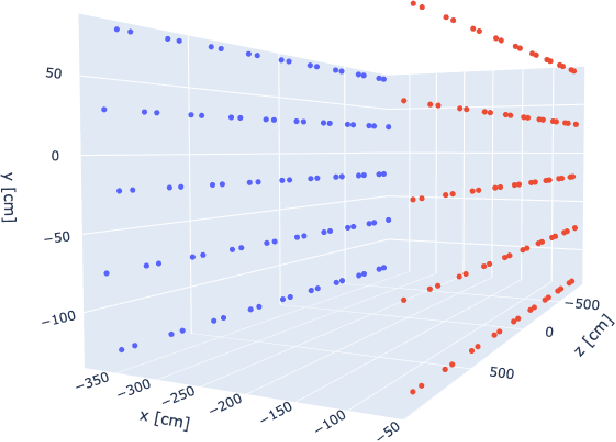
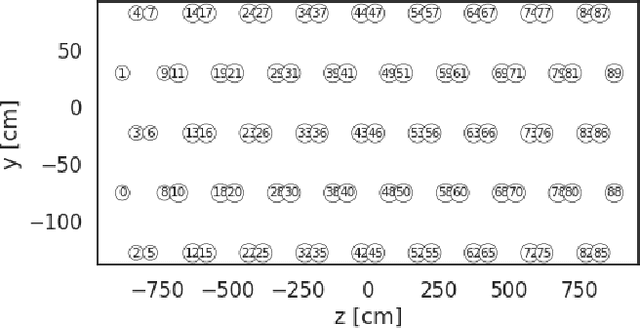
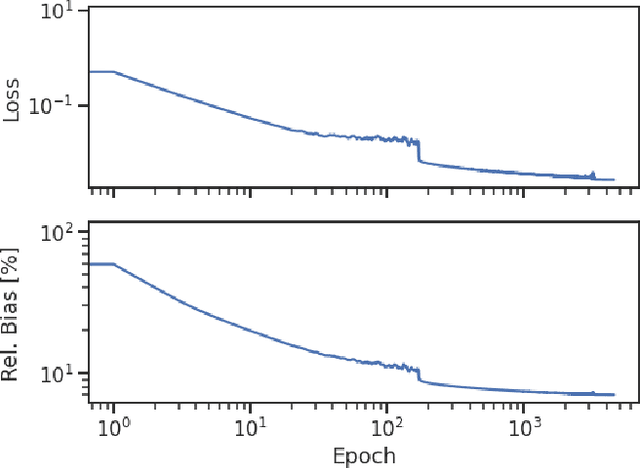
Abstract:Optical photons are used as signal in a wide variety of particle detectors. Modern neutrino experiments employ hundreds to tens of thousands of photon detectors to observe signal from millions to billions of scintillation photons produced from energy deposition of charged particles. These neutrino detectors are typically large, containing kilotons of target volume, with different optical properties. Modeling individual photon propagation in form of look-up table requires huge computational resources. As the size of a table increases with detector volume for a fixed resolution, this method scales poorly for future larger detectors. Alternative approaches such as fitting a polynomial to the model could address the memory issue, but results in poorer performance. Both look-up table and fitting approaches are prone to discrepancies between the detector simulation and the data collected. We propose a new approach using SIREN, an implicit neural representation with periodic activation functions, to model the look-up table as a 3D scene and reproduces the acceptance map with high accuracy. The number of parameters in our SIREN model is orders of magnitude smaller than the number of voxels in the look-up table. As it models an underlying functional shape, SIREN is scalable to a larger detector. Furthermore, SIREN can successfully learn the spatial gradients of the photon library, providing additional information for downstream applications. Finally, as SIREN is a neural network representation, it is differentiable with respect to its parameters, and therefore tunable via gradient descent. We demonstrate the potential of optimizing SIREN directly on real data, which mitigates the concern of data vs. simulation discrepancies. We further present an application for data reconstruction where SIREN is used to form a likelihood function for photon statistics.
Heterogeneous reconstruction of deformable atomic models in Cryo-EM
Sep 29, 2022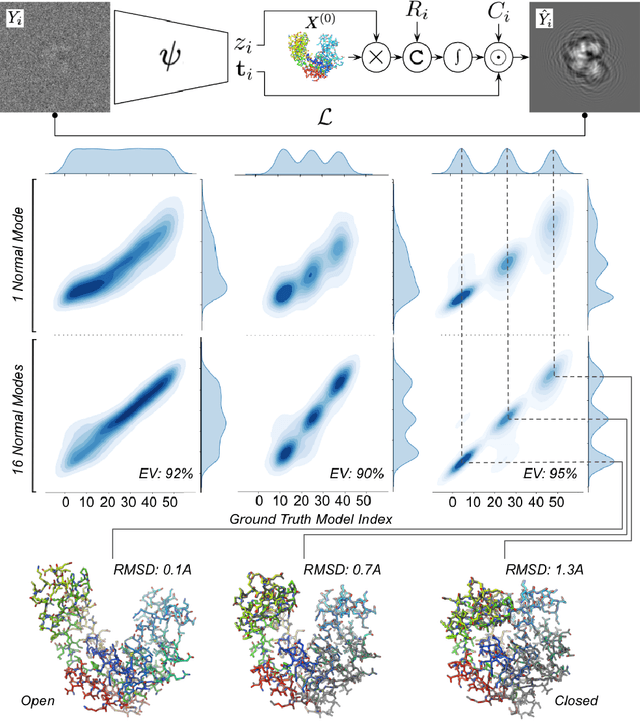
Abstract:Cryogenic electron microscopy (cryo-EM) provides a unique opportunity to study the structural heterogeneity of biomolecules. Being able to explain this heterogeneity with atomic models would help our understanding of their functional mechanisms but the size and ruggedness of the structural space (the space of atomic 3D cartesian coordinates) presents an immense challenge. Here, we describe a heterogeneous reconstruction method based on an atomistic representation whose deformation is reduced to a handful of collective motions through normal mode analysis. Our implementation uses an autoencoder. The encoder jointly estimates the amplitude of motion along the normal modes and the 2D shift between the center of the image and the center of the molecule . The physics-based decoder aggregates a representation of the heterogeneity readily interpretable at the atomic level. We illustrate our method on 3 synthetic datasets corresponding to different distributions along a simulated trajectory of adenylate kinase transitioning from its open to its closed structures. We show for each distribution that our approach is able to recapitulate the intermediate atomic models with atomic-level accuracy.
CryoAI: Amortized Inference of Poses for Ab Initio Reconstruction of 3D Molecular Volumes from Real Cryo-EM Images
Mar 16, 2022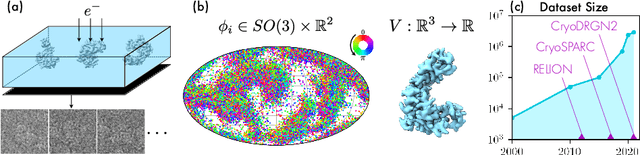
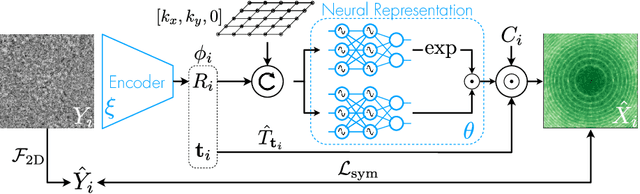
Abstract:Cryo-electron microscopy (cryo-EM) has become a tool of fundamental importance in structural biology, helping us understand the basic building blocks of life. The algorithmic challenge of cryo-EM is to jointly estimate the unknown 3D poses and the 3D electron scattering potential of a biomolecule from millions of extremely noisy 2D images. Existing reconstruction algorithms, however, cannot easily keep pace with the rapidly growing size of cryo-EM datasets due to their high computational and memory cost. We introduce cryoAI, an ab initio reconstruction algorithm for homogeneous conformations that uses direct gradient-based optimization of particle poses and the electron scattering potential from single-particle cryo-EM data. CryoAI combines a learned encoder that predicts the poses of each particle image with a physics-based decoder to aggregate each particle image into an implicit representation of the scattering potential volume. This volume is stored in the Fourier domain for computational efficiency and leverages a modern coordinate network architecture for memory efficiency. Combined with a symmetrized loss function, this framework achieves results of a quality on par with state-of-the-art cryo-EM solvers for both simulated and experimental data, one order of magnitude faster for large datasets and with significantly lower memory requirements than existing methods.
Ptychopy: GPU framework for ptychographic data analysis
Jan 24, 2022Abstract:X-ray ptychography imaging at synchrotron facilities like the Advanced Photon Source (APS) involves controlling instrument hardwares to collect a set of diffraction patterns from overlapping coherent illumination spots on extended samples, managing data storage, reconstructing ptychographic images from acquired diffraction patterns, and providing the visualization of results and feedback. In addition to the complicated workflow, ptychography instrument could produce up to several TB's of data per second that is needed to be processed in real time. This brings up the need to develop a high performance, robust and user friendly processing software package for ptychographic data analysis. In this paper we present a software framework which provides functionality of visualization, work flow control, and data reconstruction. To accelerate the computation and large datasets process, the data reconstruction part is implemented with three algorithms, ePIE, DM and LSQML using CUDA-C on GPU.
Real-time sparse-sampled Ptychographic imaging through deep neural networks
Apr 15, 2020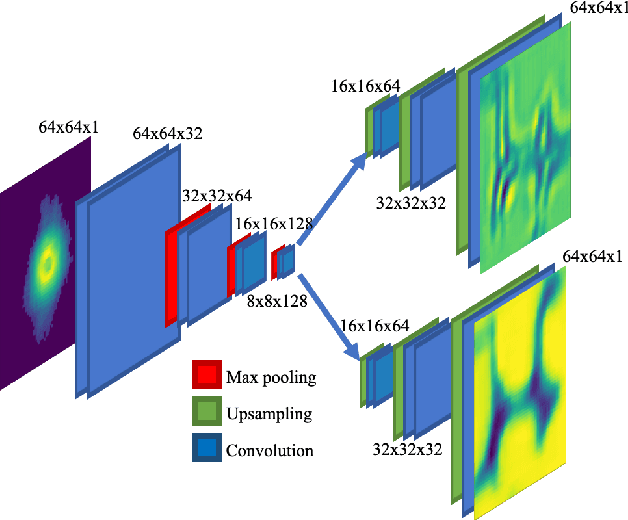
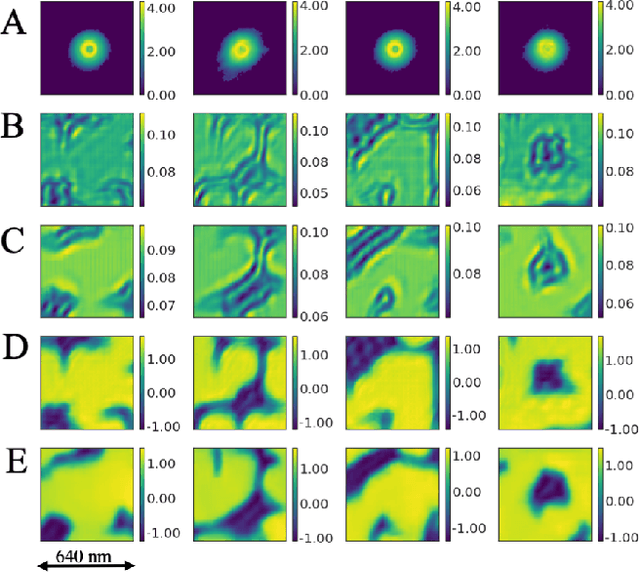
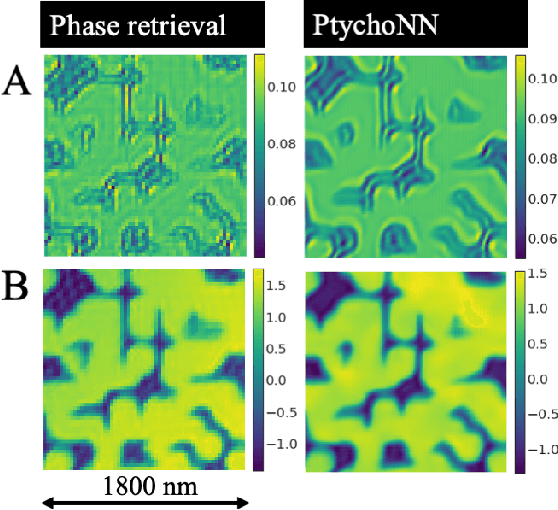
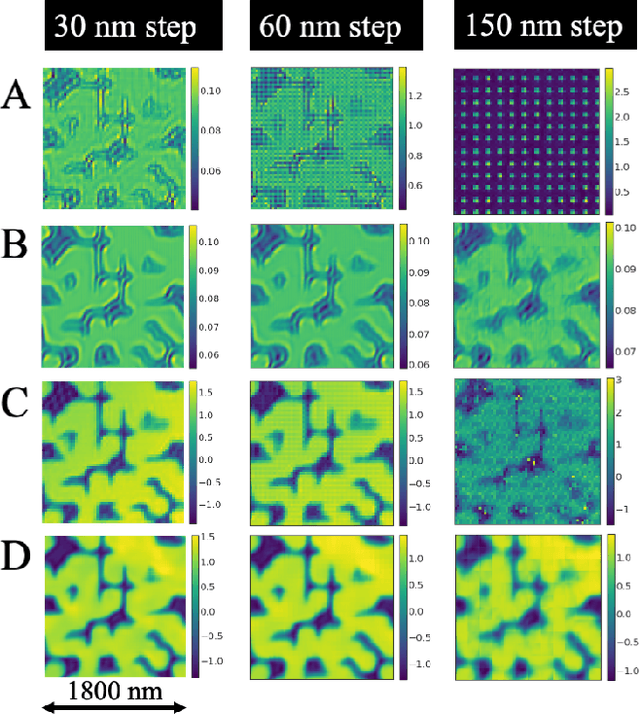
Abstract:Ptychography has rapidly grown in the fields of X-ray and electron imaging for its unprecedented ability to achieve nano or atomic scale resolution while simultaneously retrieving chemical or magnetic information from a sample. A ptychographic reconstruction is achieved by means of solving a complex inverse problem that imposes constraints both on the acquisition and on the analysis of the data, which typically precludes real-time imaging due to computational cost involved in solving this inverse problem. In this work we propose PtychoNN, a novel approach to solve the ptychography reconstruction problem based on deep convolutional neural networks. We demonstrate how the proposed method can be used to predict real-space structure and phase at each scan point solely from the corresponding far-field diffraction data. The presented results demonstrate how PtychoNN can effectively be used on experimental data, being able to generate high quality reconstructions of a sample up to hundreds of times faster than state-of-the-art ptychography reconstruction solutions once trained. By surpassing the typical constraints of iterative model-based methods, we can significantly relax the data acquisition sampling conditions and produce equally satisfactory reconstructions. Besides drastically accelerating acquisition and analysis, this capability can enable new imaging scenarios that were not possible before, in cases of dose sensitive, dynamic and extremely voluminous samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge