Retinal Vessel Segmentation
Retinal vessel segmentation is the process of identifying and segmenting blood vessels in retinal images for medical diagnosis.
Papers and Code
TFFM: Topology-Aware Feature Fusion Module via Latent Graph Reasoning for Retinal Vessel Segmentation
Jan 27, 2026Precise segmentation of retinal arteries and veins carries the diagnosis of systemic cardiovascular conditions. However, standard convolutional architectures often yield topologically disjointed segmentations, characterized by gaps and discontinuities that render reliable graph-based clinical analysis impossible despite high pixel-level accuracy. To address this, we introduce a topology-aware framework engineered to maintain vascular connectivity. Our architecture fuses a Topological Feature Fusion Module (TFFM) that maps local feature representations into a latent graph space, deploying Graph Attention Networks to capture global structural dependencies often missed by fixed receptive fields. Furthermore, we drive the learning process with a hybrid objective function, coupling Tversky loss for class imbalance with soft clDice loss to explicitly penalize topological disconnects. Evaluation on the Fundus-AVSeg dataset reveals state-of-the-art performance, achieving a combined Dice score of 90.97% and a 95% Hausdorff Distance of 3.50 pixels. Notably, our method decreases vessel fragmentation by approximately 38% relative to baselines, yielding topologically coherent vascular trees viable for automated biomarker quantification. We open-source our code at https://tffm-module.github.io/.
MTFlow: Time-Conditioned Flow Matching for Microtubule Segmentation in Noisy Microscopy Images
Jan 21, 2026Microtubules are cytoskeletal filaments that play essential roles in many cellular processes and are key therapeutic targets in several diseases. Accurate segmentation of microtubule networks is critical for studying their organization and dynamics but remains challenging due to filament curvature, dense crossings, and image noise. We present MTFlow, a novel time-conditioned flow-matching model for microtubule segmentation. Unlike conventional U-Net variants that predict masks in a single pass, MTFlow learns vector fields that iteratively transport noisy masks toward the ground truth, enabling interpretable, trajectory-based refinement. Our architecture combines a U-Net backbone with temporal embeddings, allowing the model to capture the dynamics of uncertainty resolution along filament boundaries. We trained and evaluated MTFlow on synthetic and real microtubule datasets and assessed its generalization capability on public biomedical datasets of curvilinear structures such as retinal blood vessels and nerves. MTFlow achieves competitive segmentation accuracy comparable to state-of-the-art models, offering a powerful and time-efficient tool for filamentous structure analysis with more precise annotations than manual or semi-automatic approaches.
WaveRNet: Wavelet-Guided Frequency Learning for Multi-Source Domain-Generalized Retinal Vessel Segmentation
Jan 09, 2026Domain-generalized retinal vessel segmentation is critical for automated ophthalmic diagnosis, yet faces significant challenges from domain shift induced by non-uniform illumination and varying contrast, compounded by the difficulty of preserving fine vessel structures. While the Segment Anything Model (SAM) exhibits remarkable zero-shot capabilities, existing SAM-based methods rely on simple adapter fine-tuning while overlooking frequency-domain information that encodes domain-invariant features, resulting in degraded generalization under illumination and contrast variations. Furthermore, SAM's direct upsampling inevitably loses fine vessel details. To address these limitations, we propose WaveRNet, a wavelet-guided frequency learning framework for robust multi-source domain-generalized retinal vessel segmentation. Specifically, we devise a Spectral-guided Domain Modulator (SDM) that integrates wavelet decomposition with learnable domain tokens, enabling the separation of illumination-robust low-frequency structures from high-frequency vessel boundaries while facilitating domain-specific feature generation. Furthermore, we introduce a Frequency-Adaptive Domain Fusion (FADF) module that performs intelligent test-time domain selection through wavelet-based frequency similarity and soft-weighted fusion. Finally, we present a Hierarchical Mask-Prompt Refiner (HMPR) that overcomes SAM's upsampling limitation through coarse-to-fine refinement with long-range dependency modeling. Extensive experiments under the Leave-One-Domain-Out protocol on four public retinal datasets demonstrate that WaveRNet achieves state-of-the-art generalization performance. The source code is available at https://github.com/Chanchan-Wang/WaveRNet.
TopoLoRA-SAM: Topology-Aware Parameter-Efficient Adaptation of Foundation Segmenters for Thin-Structure and Cross-Domain Binary Semantic Segmentation
Jan 05, 2026Foundation segmentation models such as the Segment Anything Model (SAM) exhibit strong zero-shot generalization through large-scale pretraining, but adapting them to domain-specific semantic segmentation remains challenging, particularly for thin structures (e.g., retinal vessels) and noisy modalities (e.g., SAR imagery). Full fine-tuning is computationally expensive and risks catastrophic forgetting. We propose \textbf{TopoLoRA-SAM}, a topology-aware and parameter-efficient adaptation framework for binary semantic segmentation. TopoLoRA-SAM injects Low-Rank Adaptation (LoRA) into the frozen ViT encoder, augmented with a lightweight spatial convolutional adapter and optional topology-aware supervision via differentiable clDice. We evaluate our approach on five benchmarks spanning retinal vessel segmentation (DRIVE, STARE, CHASE\_DB1), polyp segmentation (Kvasir-SEG), and SAR sea/land segmentation (SL-SSDD), comparing against U-Net, DeepLabV3+, SegFormer, and Mask2Former. TopoLoRA-SAM achieves the best retina-average Dice and the best overall average Dice across datasets, while training only \textbf{5.2\%} of model parameters ($\sim$4.9M). On the challenging CHASE\_DB1 dataset, our method substantially improves segmentation accuracy and robustness, demonstrating that topology-aware parameter-efficient adaptation can match or exceed fully fine-tuned specialist models. Code is available at : https://github.com/salimkhazem/Seglab.git
Adaptive Frequency Domain Alignment Network for Medical image segmentation
Dec 20, 2025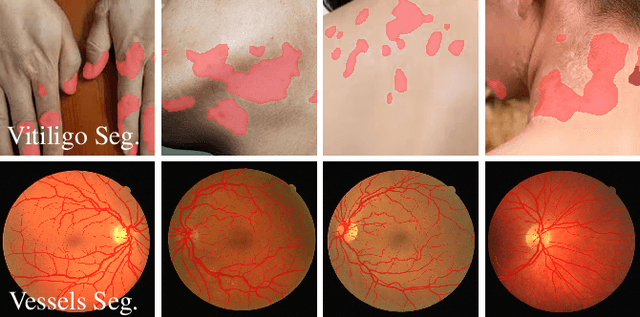
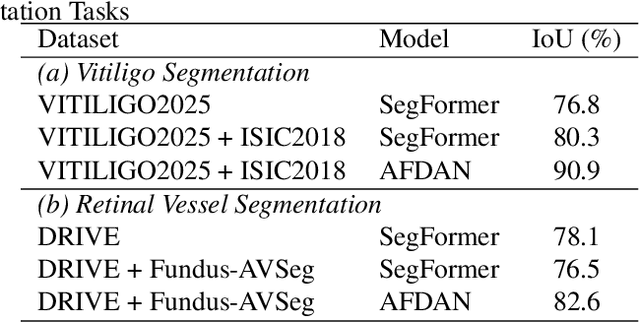
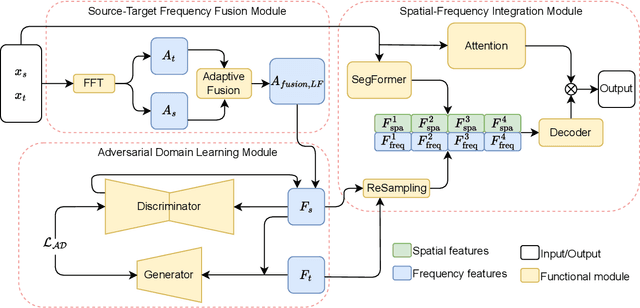
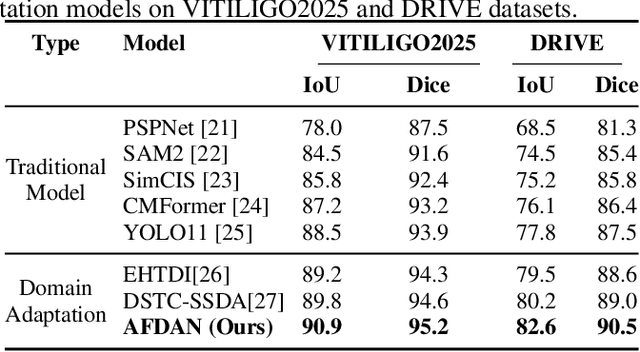
High-quality annotated data plays a crucial role in achieving accurate segmentation. However, such data for medical image segmentation are often scarce due to the time-consuming and labor-intensive nature of manual annotation. To address this challenge, we propose the Adaptive Frequency Domain Alignment Network (AFDAN)--a novel domain adaptation framework designed to align features in the frequency domain and alleviate data scarcity. AFDAN integrates three core components to enable robust cross-domain knowledge transfer: an Adversarial Domain Learning Module that transfers features from the source to the target domain; a Source-Target Frequency Fusion Module that blends frequency representations across domains; and a Spatial-Frequency Integration Module that combines both frequency and spatial features to further enhance segmentation accuracy across domains. Extensive experiments demonstrate the effectiveness of AFDAN: it achieves an Intersection over Union (IoU) of 90.9% for vitiligo segmentation in the newly constructed VITILIGO2025 dataset and a competitive IoU of 82.6% on the retinal vessel segmentation benchmark DRIVE, surpassing existing state-of-the-art approaches.
Robust Multi-Disease Retinal Classification via Xception-Based Transfer Learning and W-Net Vessel Segmentation
Dec 11, 2025In recent years, the incidence of vision-threatening eye diseases has risen dramatically, necessitating scalable and accurate screening solutions. This paper presents a comprehensive study on deep learning architectures for the automated diagnosis of ocular conditions. To mitigate the "black-box" limitations of standard convolutional neural networks (CNNs), we implement a pipeline that combines deep feature extraction with interpretable image processing modules. Specifically, we focus on high-fidelity retinal vessel segmentation as an auxiliary task to guide the classification process. By grounding the model's predictions in clinically relevant morphological features, we aim to bridge the gap between algorithmic output and expert medical validation, thereby reducing false positives and improving deployment viability in clinical settings.
rNCA: Self-Repairing Segmentation Masks
Dec 15, 2025Accurately predicting topologically correct masks remains a difficult task for general segmentation models, which often produce fragmented or disconnected outputs. Fixing these artifacts typically requires hand-crafted refinement rules or architectures specialized to a particular task. Here, we show that Neural Cellular Automata (NCA) can be directly re-purposed as an effective refinement mechanism, using local, iterative updates guided by image context to repair segmentation masks. By training on imperfect masks and ground truths, the automaton learns the structural properties of the target shape while relying solely on local information. When applied to coarse, globally predicted masks, the learned dynamics progressively reconnect broken regions, prune loose fragments and converge towards stable, topologically consistent results. We show how refinement NCA (rNCA) can be easily applied to repair common topological errors produced by different base segmentation models and tasks: for fragmented retinal vessels, it yields 2-3% gains in Dice/clDice and improves Betti errors, reducing $β_0$ errors by 60% and $β_1$ by 20%; for myocardium, it repairs 61.5% of broken cases in a zero-shot setting while lowering ASSD and HD by 19% and 16%, respectively. This showcases NCA as effective and broadly applicable refiners.
Rotterdam artery-vein segmentation (RAV) dataset
Dec 19, 2025Purpose: To provide a diverse, high-quality dataset of color fundus images (CFIs) with detailed artery-vein (A/V) segmentation annotations, supporting the development and evaluation of machine learning algorithms for vascular analysis in ophthalmology. Methods: CFIs were sampled from the longitudinal Rotterdam Study (RS), encompassing a wide range of ages, devices, and capture conditions. Images were annotated using a custom interface that allowed graders to label arteries, veins, and unknown vessels on separate layers, starting from an initial vessel segmentation mask. Connectivity was explicitly verified and corrected using connected component visualization tools. Results: The dataset includes 1024x1024-pixel PNG images in three modalities: original RGB fundus images, contrast-enhanced versions, and RGB-encoded A/V masks. Image quality varied widely, including challenging samples typically excluded by automated quality assessment systems, but judged to contain valuable vascular information. Conclusion: This dataset offers a rich and heterogeneous source of CFIs with high-quality segmentations. It supports robust benchmarking and training of machine learning models under real-world variability in image quality and acquisition settings. Translational Relevance: By including connectivity-validated A/V masks and diverse image conditions, this dataset enables the development of clinically applicable, generalizable machine learning tools for retinal vascular analysis, potentially improving automated screening and diagnosis of systemic and ocular diseases.
XOCT: Enhancing OCT to OCTA Translation via Cross-Dimensional Supervised Multi-Scale Feature Learning
Sep 09, 2025Optical Coherence Tomography Angiography (OCTA) and its derived en-face projections provide high-resolution visualization of the retinal and choroidal vasculature, which is critical for the rapid and accurate diagnosis of retinal diseases. However, acquiring high-quality OCTA images is challenging due to motion sensitivity and the high costs associated with software modifications for conventional OCT devices. Moreover, current deep learning methods for OCT-to-OCTA translation often overlook the vascular differences across retinal layers and struggle to reconstruct the intricate, dense vascular details necessary for reliable diagnosis. To overcome these limitations, we propose XOCT, a novel deep learning framework that integrates Cross-Dimensional Supervision (CDS) with a Multi-Scale Feature Fusion (MSFF) network for layer-aware vascular reconstruction. Our CDS module leverages 2D layer-wise en-face projections, generated via segmentation-weighted z-axis averaging, as supervisory signals to compel the network to learn distinct representations for each retinal layer through fine-grained, targeted guidance. Meanwhile, the MSFF module enhances vessel delineation through multi-scale feature extraction combined with a channel reweighting strategy, effectively capturing vascular details at multiple spatial scales. Our experiments on the OCTA-500 dataset demonstrate XOCT's improvements, especially for the en-face projections which are significant for clinical evaluation of retinal pathologies, underscoring its potential to enhance OCTA accessibility, reliability, and diagnostic value for ophthalmic disease detection and monitoring. The code is available at https://github.com/uci-cbcl/XOCT.
Adaptive Attention Residual U-Net for curvilinear structure segmentation in fluorescence microscopy and biomedical images
Jul 10, 2025Segmenting curvilinear structures in fluorescence microscopy remains a challenging task, particularly under noisy conditions and in dense filament networks commonly seen in vivo. To address this, we created two original datasets consisting of hundreds of synthetic images of fluorescently labelled microtubules within cells. These datasets are precisely annotated and closely mimic real microscopy images, including realistic noise. The second dataset presents an additional challenge, by simulating varying fluorescence intensities along filaments that complicate segmentation. While deep learning has shown strong potential in biomedical image analysis, its performance often declines in noisy or low-contrast conditions. To overcome this limitation, we developed a novel advanced architecture: the Adaptive Squeeze-and-Excitation Residual U-Net (ASE_Res_UNet). This model enhanced the standard U-Net by integrating residual blocks in the encoder and adaptive SE attention mechanisms in the decoder. Through ablation studies and comprehensive visual and quantitative evaluations, ASE_Res_UNet consistently outperformed its variants, namely standard U-Net, ASE_UNet and Res_UNet architectures. These improvements, particularly in noise resilience and detecting fine, low-intensity structures, were largely attributed to the adaptive SE attention module that we created. We further benchmarked ASE_Res_UNet against various state-of-the-art models, and found it achieved superior performance on our most challenging dataset. Finally, the model also generalized well to real microscopy images of stained microtubules as well as to other curvilinear structures. Indeed, it successfully segmented retinal blood vessels and nerves in noisy or low-contrast biomedical images, demonstrating its strong potential for applications in disease diagnosis and treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge