Pengfei Shao
Alzheimer's Disease Neuroimaging Initiative, the Australian Imaging Biomarkers and Lifestyle flagship study of ageing
Intermediate Domain-guided Adaptation for Unsupervised Chorioallantoic Membrane Vessel Segmentation
Mar 06, 2025



Abstract:The chorioallantoic membrane (CAM) model is widely employed in angiogenesis research, and distribution of growing blood vessels is the key evaluation indicator. As a result, vessel segmentation is crucial for quantitative assessment based on topology and morphology. However, manual segmentation is extremely time-consuming, labor-intensive, and prone to inconsistency due to its subjective nature. Moreover, research on CAM vessel segmentation algorithms remains limited, and the lack of public datasets contributes to poor prediction performance. To address these challenges, we propose an innovative Intermediate Domain-guided Adaptation (IDA) method, which utilizes the similarity between CAM images and retinal images, along with existing public retinal datasets, to perform unsupervised training on CAM images. Specifically, we introduce a Multi-Resolution Asymmetric Translation (MRAT) strategy to generate intermediate images to promote image-level interaction. Then, an Intermediate Domain-guided Contrastive Learning (IDCL) module is developed to disentangle cross-domain feature representations. This method overcomes the limitations of existing unsupervised domain adaptation (UDA) approaches, which primarily concentrate on directly source-target alignment while neglecting intermediate domain information. Notably, we create the first CAM dataset to validate the proposed algorithm. Extensive experiments on this dataset show that our method outperforms compared approaches. Moreover, it achieves superior performance in UDA tasks across retinal datasets, highlighting its strong generalization capability. The CAM dataset and source codes are available at https://github.com/Light-47/IDA.
MCPA: Multi-scale Cross Perceptron Attention Network for 2D Medical Image Segmentation
Jul 27, 2023Abstract:The UNet architecture, based on Convolutional Neural Networks (CNN), has demonstrated its remarkable performance in medical image analysis. However, it faces challenges in capturing long-range dependencies due to the limited receptive fields and inherent bias of convolutional operations. Recently, numerous transformer-based techniques have been incorporated into the UNet architecture to overcome this limitation by effectively capturing global feature correlations. However, the integration of the Transformer modules may result in the loss of local contextual information during the global feature fusion process. To overcome these challenges, we propose a 2D medical image segmentation model called Multi-scale Cross Perceptron Attention Network (MCPA). The MCPA consists of three main components: an encoder, a decoder, and a Cross Perceptron. The Cross Perceptron first captures the local correlations using multiple Multi-scale Cross Perceptron modules, facilitating the fusion of features across scales. The resulting multi-scale feature vectors are then spatially unfolded, concatenated, and fed through a Global Perceptron module to model global dependencies. Furthermore, we introduce a Progressive Dual-branch Structure to address the semantic segmentation of the image involving finer tissue structures. This structure gradually shifts the segmentation focus of MCPA network training from large-scale structural features to more sophisticated pixel-level features. We evaluate our proposed MCPA model on several publicly available medical image datasets from different tasks and devices, including the open large-scale dataset of CT (Synapse), MRI (ACDC), fundus camera (DRIVE, CHASE_DB1, HRF), and OCTA (ROSE). The experimental results show that our MCPA model achieves state-of-the-art performance. The code is available at https://github.com/simonustc/MCPA-for-2D-Medical-Image-Segmentation.
Phased Progressive Learning with Coupling-Regulation-Imbalance Loss for Imbalanced Classification
May 24, 2022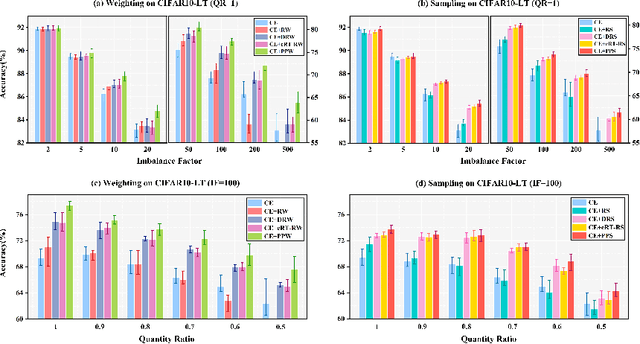
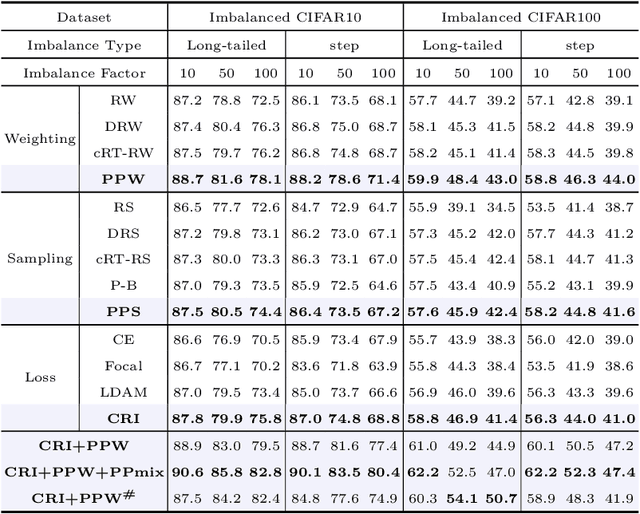
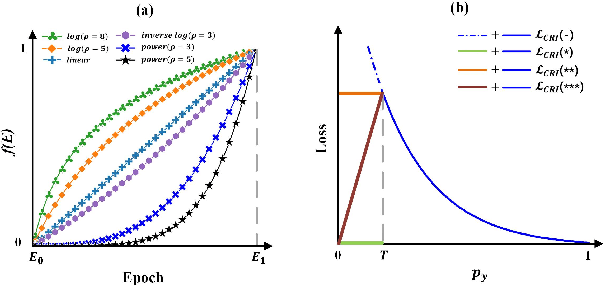
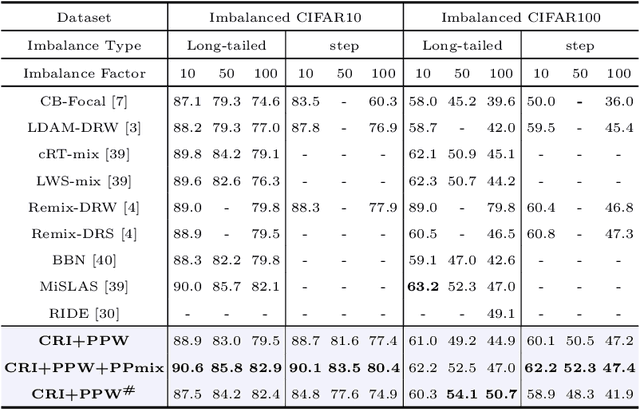
Abstract:Deep neural networks generally perform poorly with datasets that suffer from quantity imbalance and classification difficulty imbalance between different classes. In order to alleviate the problem of dataset bias or domain shift in the existing two-stage approaches, a phased progressive learning schedule was proposed for smoothly transferring the training emphasis from representation learning to upper classifier training. This has greater effectivity on datasets that have more severe imbalances or smaller scales. A coupling-regulation-imbalance loss function was designed, coupling a correction term, Focal loss and LDAM loss. Coupling-regulation-imbalance loss can better deal with quantity imbalance and outliers, while regulating focus-of-attention of samples with a variety of classification difficulties. Excellent results were achieved on multiple benchmark datasets using these approaches and they can be easily generalized for other imbalanced classification models. Our code will be open source soon.
CPNet: Cycle Prototype Network for Weakly-supervised 3D Renal Compartments Segmentation on CT Images
Aug 15, 2021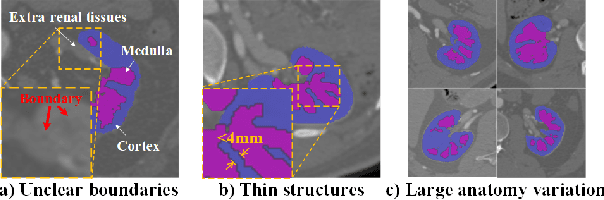
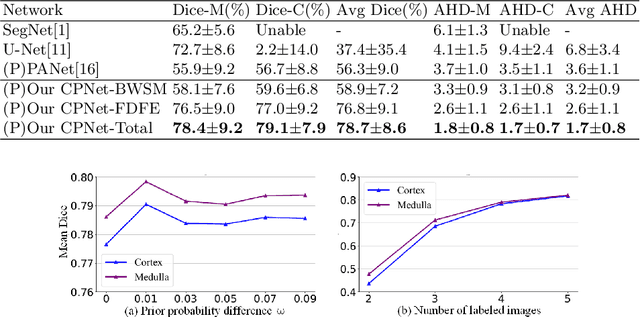
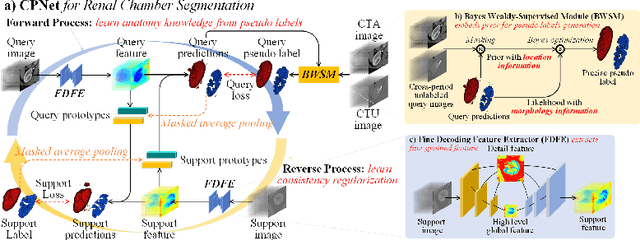
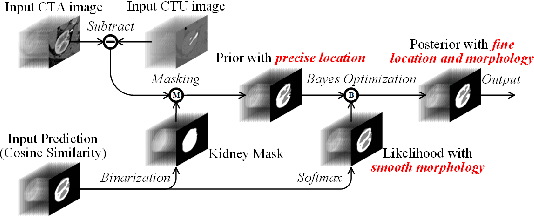
Abstract:Renal compartment segmentation on CT images targets on extracting the 3D structure of renal compartments from abdominal CTA images and is of great significance to the diagnosis and treatment for kidney diseases. However, due to the unclear compartment boundary, thin compartment structure and large anatomy variation of 3D kidney CT images, deep-learning based renal compartment segmentation is a challenging task. We propose a novel weakly supervised learning framework, Cycle Prototype Network, for 3D renal compartment segmentation. It has three innovations: 1) A Cycle Prototype Learning (CPL) is proposed to learn consistency for generalization. It learns from pseudo labels through the forward process and learns consistency regularization through the reverse process. The two processes make the model robust to noise and label-efficient. 2) We propose a Bayes Weakly Supervised Module (BWSM) based on cross-period prior knowledge. It learns prior knowledge from cross-period unlabeled data and perform error correction automatically, thus generates accurate pseudo labels. 3) We present a Fine Decoding Feature Extractor (FDFE) for fine-grained feature extraction. It combines global morphology information and local detail information to obtain feature maps with sharp detail, so the model will achieve fine segmentation on thin structures. Our model achieves Dice of 79.1% and 78.7% with only four labeled images, achieving a significant improvement by about 20% than typical prototype model PANet.
An explainable two-dimensional single model deep learning approach for Alzheimer's disease diagnosis and brain atrophy localization
Jul 28, 2021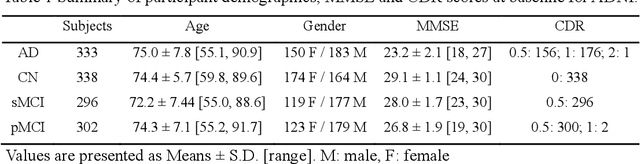
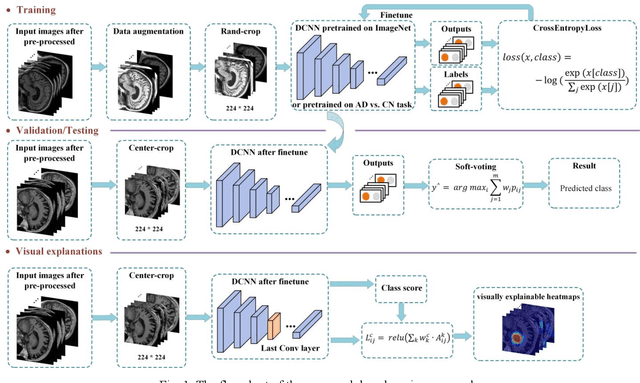
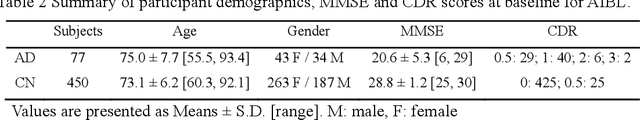
Abstract:Early and accurate diagnosis of Alzheimer's disease (AD) and its prodromal period mild cognitive impairment (MCI) is essential for the delayed disease progression and the improved quality of patients'life. The emerging computer-aided diagnostic methods that combine deep learning with structural magnetic resonance imaging (sMRI) have achieved encouraging results, but some of them are limit of issues such as data leakage and unexplainable diagnosis. In this research, we propose a novel end-to-end deep learning approach for automated diagnosis of AD and localization of important brain regions related to the disease from sMRI data. This approach is based on a 2D single model strategy and has the following differences from the current approaches: 1) Convolutional Neural Network (CNN) models of different structures and capacities are evaluated systemically and the most suitable model is adopted for AD diagnosis; 2) a data augmentation strategy named Two-stage Random RandAugment (TRRA) is proposed to alleviate the overfitting issue caused by limited training data and to improve the classification performance in AD diagnosis; 3) an explainable method of Grad-CAM++ is introduced to generate the visually explainable heatmaps that localize and highlight the brain regions that our model focuses on and to make our model more transparent. Our approach has been evaluated on two publicly accessible datasets for two classification tasks of AD vs. cognitively normal (CN) and progressive MCI (pMCI) vs. stable MCI (sMCI). The experimental results indicate that our approach outperforms the state-of-the-art approaches, including those using multi-model and 3D CNN methods. The resultant localization heatmaps from our approach also highlight the lateral ventricle and some disease-relevant regions of cortex, coincident with the commonly affected regions during the development of AD.
Single Model Deep Learning on Imbalanced Small Datasets for Skin Lesion Classification
Feb 02, 2021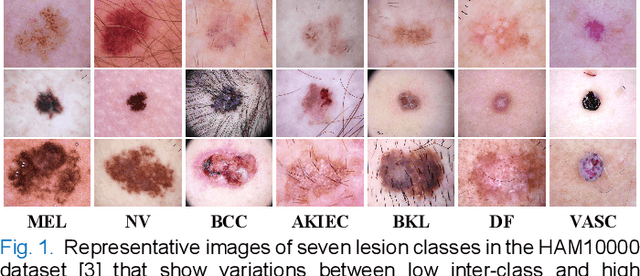
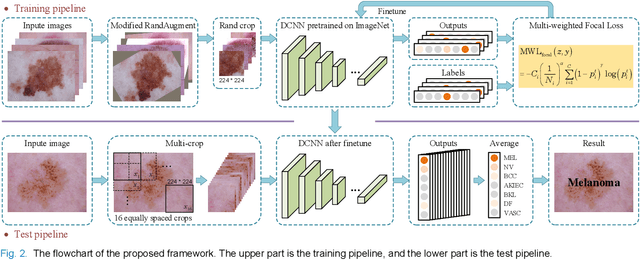
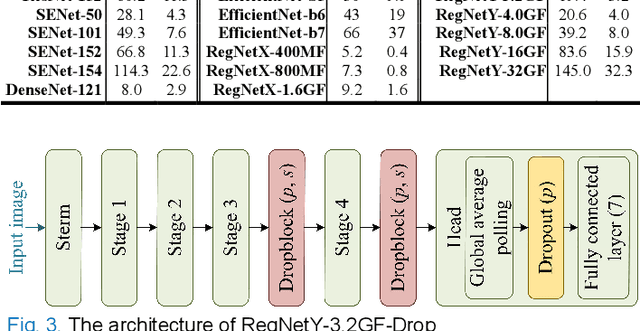
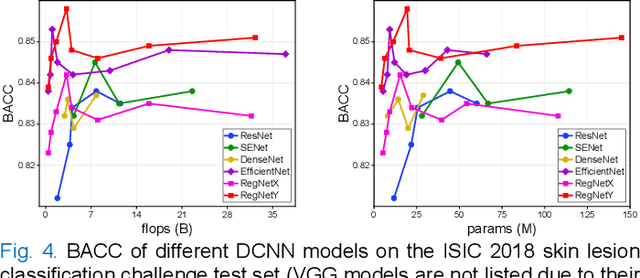
Abstract:Deep convolutional neural network (DCNN) models have been widely explored for skin disease diagnosis and some of them have achieved the diagnostic outcomes comparable or even superior to those of dermatologists. However, broad implementation of DCNN in skin disease detection is hindered by small size and data imbalance of the publically accessible skin lesion datasets. This paper proposes a novel data augmentation strategy for single model classification of skin lesions based on a small and imbalanced dataset. First, various DCNNs are trained on this dataset to show that the models with moderate complexity outperform the larger models. Second, regularization DropOut and DropBlock are added to reduce overfitting and a Modified RandAugment augmentation strategy is proposed to address the defects of sample underrepresentation in the small dataset. Finally, a novel Multi-Weighted Focal Loss function is introduced to overcome the challenge of uneven sample size and classification difficulty. By combining Modified RandAugment and Multi-weighted Focal Loss in a single DCNN model, we have achieved the classification accuracy comparable to those of multiple ensembling models on the ISIC 2018 challenge test dataset. Our study shows that this method is able to achieve a high classification performance at a low cost of computational resources and inference time, potentially suitable to implement in mobile devices for automated screening of skin lesions and many other malignancies in low resource settings.
Low-cost and high-performance data augmentation for deep-learning-based skin lesion classification
Jan 07, 2021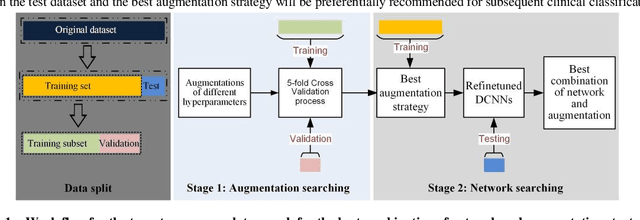
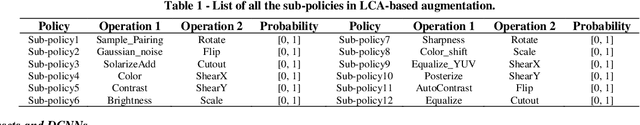
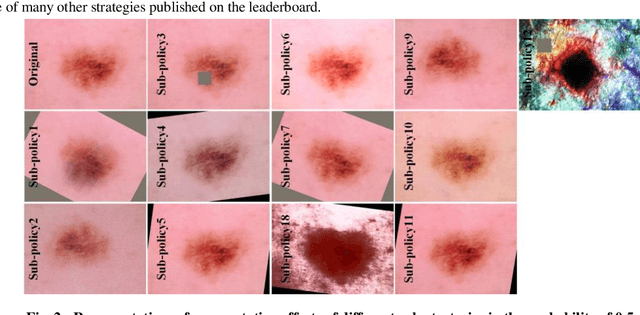
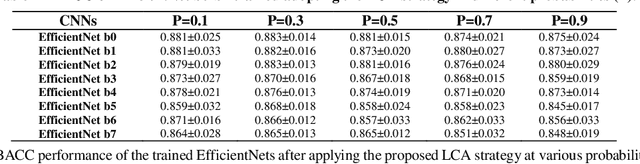
Abstract:Although deep convolutional neural networks (DCNNs) have achieved significant accuracy in skin lesion classification comparable or even superior to those of dermatologists, practical implementation of these models for skin cancer screening in low resource settings is hindered by their limitations in computational cost and training dataset. To overcome these limitations, we propose a low-cost and high-performance data augmentation strategy that includes two consecutive stages of augmentation search and network search. At the augmentation search stage, the augmentation strategy is optimized in the search space of Low-Cost-Augment (LCA) under the criteria of balanced accuracy (BACC) with 5-fold cross validation. At the network search stage, the DCNNs are fine-tuned with the full training set in order to select the model with the highest BACC. The efficiency of the proposed data augmentation strategy is verified on the HAM10000 dataset using EfficientNets as a baseline. With the proposed strategy, we are able to reduce the search space to 60 and achieve a high BACC of 0.853 by using a single DCNN model without external database, suitable to be implemented in mobile devices for DCNN-based skin lesion detection in low resource settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge