Jean-Louis Coatrieux
CRIBS, LTSI
Geometric Visual Similarity Learning in 3D Medical Image Self-supervised Pre-training
Mar 02, 2023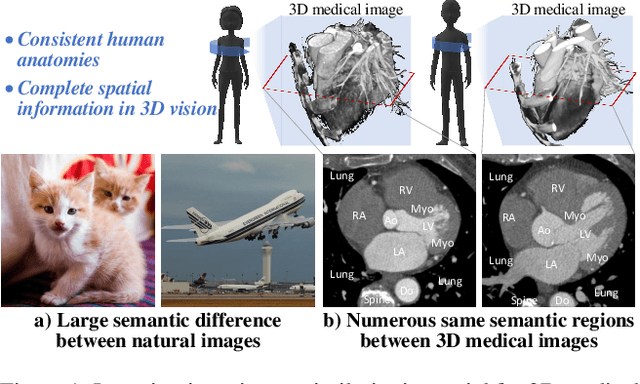
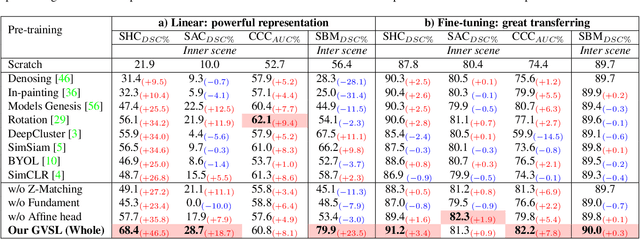
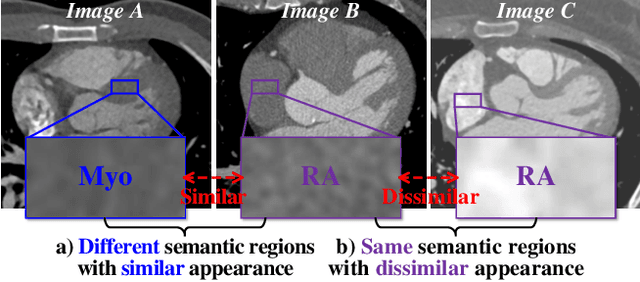

Abstract:Learning inter-image similarity is crucial for 3D medical images self-supervised pre-training, due to their sharing of numerous same semantic regions. However, the lack of the semantic prior in metrics and the semantic-independent variation in 3D medical images make it challenging to get a reliable measurement for the inter-image similarity, hindering the learning of consistent representation for same semantics. We investigate the challenging problem of this task, i.e., learning a consistent representation between images for a clustering effect of same semantic features. We propose a novel visual similarity learning paradigm, Geometric Visual Similarity Learning, which embeds the prior of topological invariance into the measurement of the inter-image similarity for consistent representation of semantic regions. To drive this paradigm, we further construct a novel geometric matching head, the Z-matching head, to collaboratively learn the global and local similarity of semantic regions, guiding the efficient representation learning for different scale-level inter-image semantic features. Our experiments demonstrate that the pre-training with our learning of inter-image similarity yields more powerful inner-scene, inter-scene, and global-local transferring ability on four challenging 3D medical image tasks. Our codes and pre-trained models will be publicly available on https://github.com/YutingHe-list/GVSL.
* Accepted by CVPR 2023
XMorpher: Full Transformer for Deformable Medical Image Registration via Cross Attention
Jun 15, 2022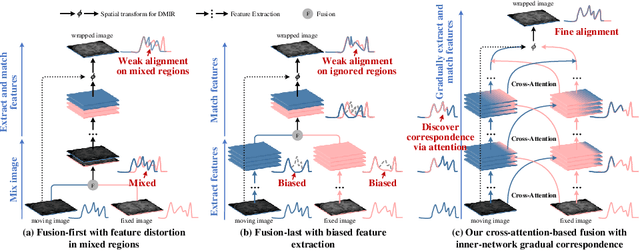
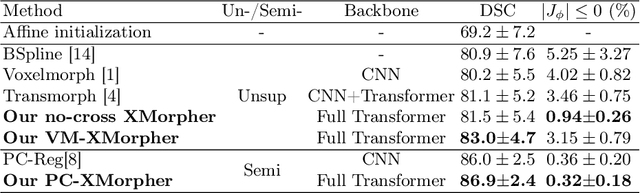
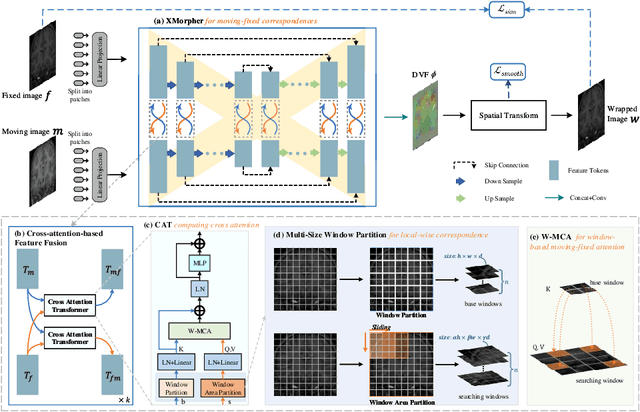
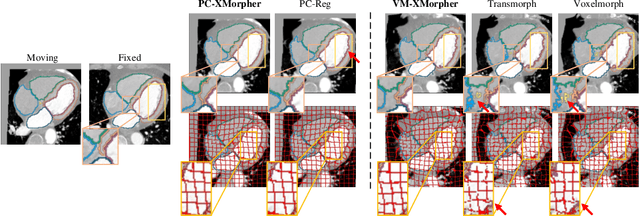
Abstract:An effective backbone network is important to deep learning-based Deformable Medical Image Registration (DMIR), because it extracts and matches the features between two images to discover the mutual correspondence for fine registration. However, the existing deep networks focus on single image situation and are limited in registration task which is performed on paired images. Therefore, we advance a novel backbone network, XMorpher, for the effective corresponding feature representation in DMIR. 1) It proposes a novel full transformer architecture including dual parallel feature extraction networks which exchange information through cross attention, thus discovering multi-level semantic correspondence while extracting respective features gradually for final effective registration. 2) It advances the Cross Attention Transformer (CAT) blocks to establish the attention mechanism between images which is able to find the correspondence automatically and prompts the features to fuse efficiently in the network. 3) It constrains the attention computation between base windows and searching windows with different sizes, and thus focuses on the local transformation of deformable registration and enhances the computing efficiency at the same time. Without any bells and whistles, our XMorpher gives Voxelmorph 2.8% improvement on DSC , demonstrating its effective representation of the features from the paired images in DMIR. We believe that our XMorpher has great application potential in more paired medical images. Our XMorpher is open on https://github.com/Solemoon/XMorpher
MNet: Rethinking 2D/3D Networks for Anisotropic Medical Image Segmentation
May 10, 2022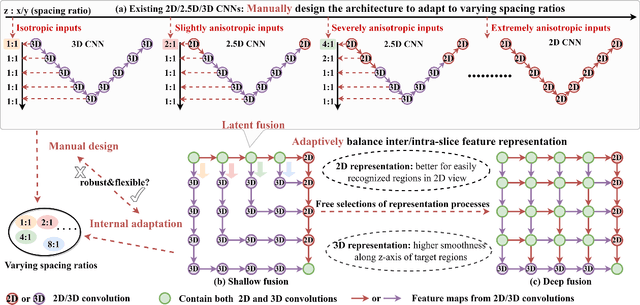
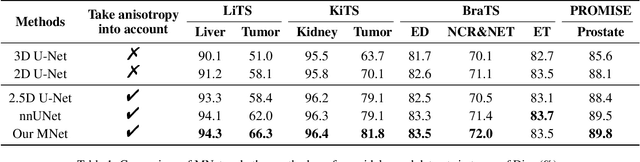
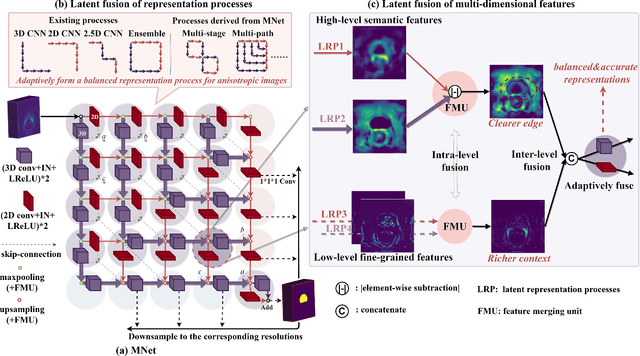

Abstract:The nature of thick-slice scanning causes severe inter-slice discontinuities of 3D medical images, and the vanilla 2D/3D convolutional neural networks (CNNs) fail to represent sparse inter-slice information and dense intra-slice information in a balanced way, leading to severe underfitting to inter-slice features (for vanilla 2D CNNs) and overfitting to noise from long-range slices (for vanilla 3D CNNs). In this work, a novel mesh network (MNet) is proposed to balance the spatial representation inter axes via learning. 1) Our MNet latently fuses plenty of representation processes by embedding multi-dimensional convolutions deeply into basic modules, making the selections of representation processes flexible, thus balancing representation for sparse inter-slice information and dense intra-slice information adaptively. 2) Our MNet latently fuses multi-dimensional features inside each basic module, simultaneously taking the advantages of 2D (high segmentation accuracy of the easily recognized regions in 2D view) and 3D (high smoothness of 3D organ contour) representations, thus obtaining more accurate modeling for target regions. Comprehensive experiments are performed on four public datasets (CT\&MR), the results consistently demonstrate the proposed MNet outperforms the other methods. The code and datasets are available at: https://github.com/zfdong-code/MNet
CPNet: Cycle Prototype Network for Weakly-supervised 3D Renal Compartments Segmentation on CT Images
Aug 15, 2021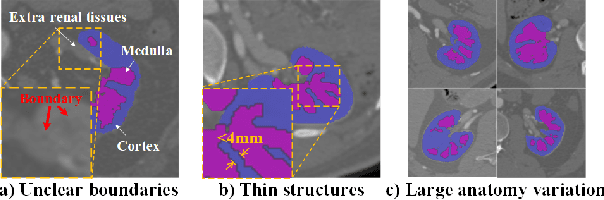
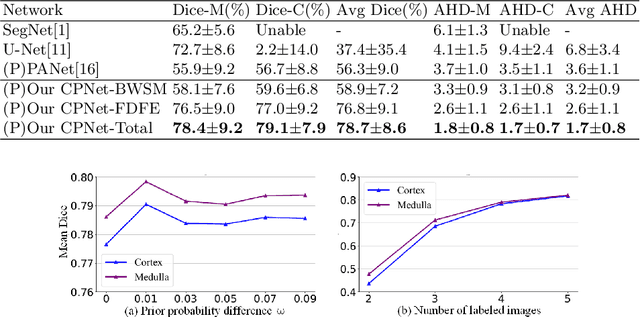
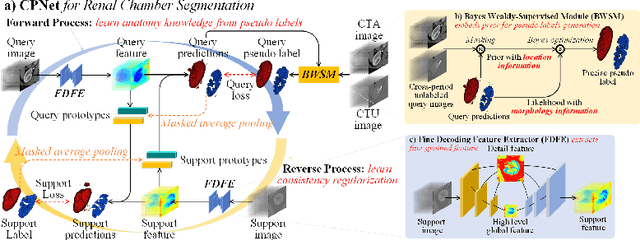
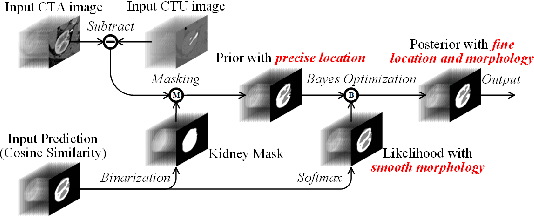
Abstract:Renal compartment segmentation on CT images targets on extracting the 3D structure of renal compartments from abdominal CTA images and is of great significance to the diagnosis and treatment for kidney diseases. However, due to the unclear compartment boundary, thin compartment structure and large anatomy variation of 3D kidney CT images, deep-learning based renal compartment segmentation is a challenging task. We propose a novel weakly supervised learning framework, Cycle Prototype Network, for 3D renal compartment segmentation. It has three innovations: 1) A Cycle Prototype Learning (CPL) is proposed to learn consistency for generalization. It learns from pseudo labels through the forward process and learns consistency regularization through the reverse process. The two processes make the model robust to noise and label-efficient. 2) We propose a Bayes Weakly Supervised Module (BWSM) based on cross-period prior knowledge. It learns prior knowledge from cross-period unlabeled data and perform error correction automatically, thus generates accurate pseudo labels. 3) We present a Fine Decoding Feature Extractor (FDFE) for fine-grained feature extraction. It combines global morphology information and local detail information to obtain feature maps with sharp detail, so the model will achieve fine segmentation on thin structures. Our model achieves Dice of 79.1% and 78.7% with only four labeled images, achieving a significant improvement by about 20% than typical prototype model PANet.
EnMcGAN: Adversarial Ensemble Learning for 3D Complete Renal Structures Segmentation
Jun 08, 2021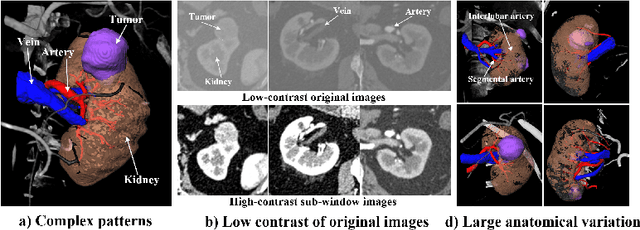
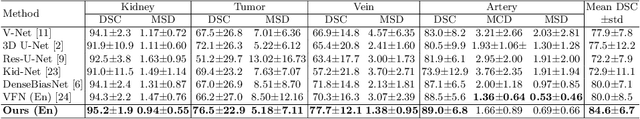
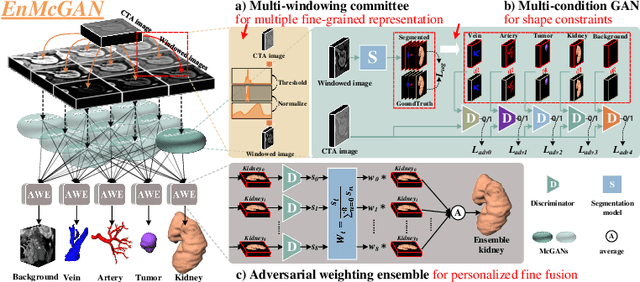
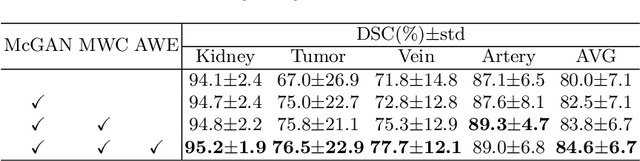
Abstract:3D complete renal structures(CRS) segmentation targets on segmenting the kidneys, tumors, renal arteries and veins in one inference. Once successful, it will provide preoperative plans and intraoperative guidance for laparoscopic partial nephrectomy(LPN), playing a key role in the renal cancer treatment. However, no success has been reported in 3D CRS segmentation due to the complex shapes of renal structures, low contrast and large anatomical variation. In this study, we utilize the adversarial ensemble learning and propose Ensemble Multi-condition GAN(EnMcGAN) for 3D CRS segmentation for the first time. Its contribution is three-fold. 1)Inspired by windowing, we propose the multi-windowing committee which divides CTA image into multiple narrow windows with different window centers and widths enhancing the contrast for salient boundaries and soft tissues. And then, it builds an ensemble segmentation model on these narrow windows to fuse the segmentation superiorities and improve whole segmentation quality. 2)We propose the multi-condition GAN which equips the segmentation model with multiple discriminators to encourage the segmented structures meeting their real shape conditions, thus improving the shape feature extraction ability. 3)We propose the adversarial weighted ensemble module which uses the trained discriminators to evaluate the quality of segmented structures, and normalizes these evaluation scores for the ensemble weights directed at the input image, thus enhancing the ensemble results. 122 patients are enrolled in this study and the mean Dice coefficient of the renal structures achieves 84.6%. Extensive experiments with promising results on renal structures reveal powerful segmentation accuracy and great clinical significance in renal cancer treatment.
Deep Complementary Joint Model for Complex Scene Registration and Few-shot Segmentation on Medical Images
Aug 03, 2020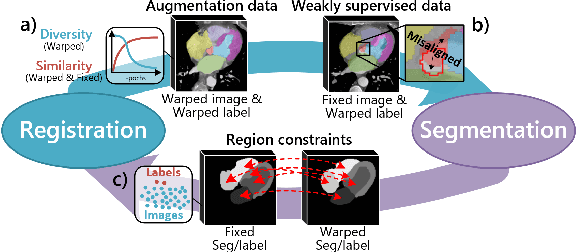
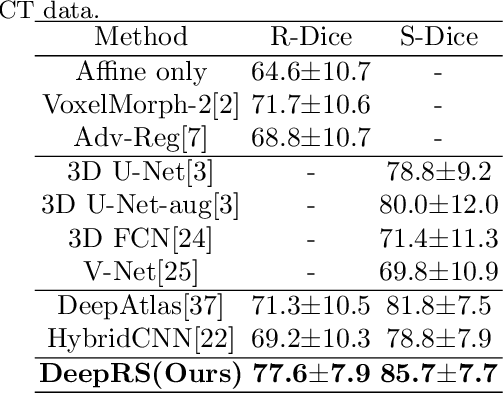
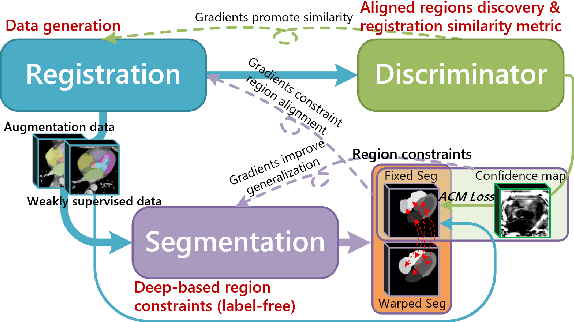
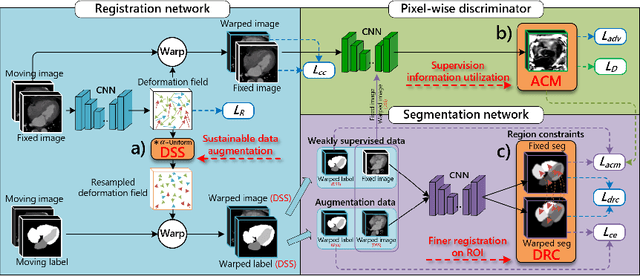
Abstract:Deep learning-based medical image registration and segmentation joint models utilize the complementarity (augmentation data or weakly supervised data from registration, region constraints from segmentation) to bring mutual improvement in complex scene and few-shot situation. However, further adoption of the joint models are hindered: 1) the diversity of augmentation data is reduced limiting the further enhancement of segmentation, 2) misaligned regions in weakly supervised data disturb the training process, 3) lack of label-based region constraints in few-shot situation limits the registration performance. We propose a novel Deep Complementary Joint Model (DeepRS) for complex scene registration and few-shot segmentation. We embed a perturbation factor in the registration to increase the activity of deformation thus maintaining the augmentation data diversity. We take a pixel-wise discriminator to extract alignment confidence maps which highlight aligned regions in weakly supervised data so the misaligned regions' disturbance will be suppressed via weighting. The outputs from segmentation model are utilized to implement deep-based region constraints thus relieving the label requirements and bringing fine registration. Extensive experiments on the CT dataset of MM-WHS 2017 Challenge show great advantages of our DeepRS that outperforms the existing state-of-the-art models.
Image reconstruction from limited range projections using orthogonal moments
Mar 12, 2014



Abstract:A set of orthonormal polynomials is proposed for image reconstruction from projection data. The relationship between the projection moments and image moments is discussed in detail, and some interesting properties are demonstrated. Simulation results are provided to validate the method and to compare its performance with previous works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge