Photoplethysmography
Photoplethysmography (PPG) is a non-invasive optical technique used to measure blood volume changes in the microvascular bed of tissue.
Papers and Code
Benchmarking and Enhancing PPG-Based Cuffless Blood Pressure Estimation Methods
Feb 04, 2026Cuffless blood pressure screening based on easily acquired photoplethysmography (PPG) signals offers a practical pathway toward scalable cardiovascular health assessment. Despite rapid progress, existing PPG-based blood pressure estimation models have not consistently achieved the established clinical numerical limits such as AAMI/ISO 81060-2, and prior evaluations often lack the rigorous experimental controls necessary for valid clinical assessment. Moreover, the publicly available datasets commonly used are heterogeneous and lack physiologically controlled conditions for fair benchmarking. To enable fair benchmarking under physiologically controlled conditions, we created a standardized benchmarking subset NBPDB comprising 101,453 high-quality PPG segments from 1,103 healthy adults, derived from MIMIC-III and VitalDB. Using this dataset, we systematically benchmarked several state-of-the-art PPG-based models. The results showed that none of the evaluated models met the AAMI/ISO 81060-2 accuracy requirements (mean error $<$ 5 mmHg and standard deviation $<$ 8 mmHg). To improve model accuracy, we modified these models and added patient demographic data such as age, sex, and body mass index as additional inputs. Our modifications consistently improved performance across all models. In particular, the MInception model reduced error by 23\% after adding the demographic data and yielded mean absolute errors of 4.75 mmHg (SBP) and 2.90 mmHg (DBP), achieves accuracy comparable to the numerical limits defined by AAMI/ISO accuracy standards. Our results show that existing PPG-based BP estimation models lack clinical practicality under standardized conditions, while incorporating demographic information markedly improves their accuracy and physiological validity.
Aortic Valve Disease Detection from PPG via Physiology-Informed Self-Supervised Learning
Feb 04, 2026Traditional diagnosis of aortic valve disease relies on echocardiography, but its cost and required expertise limit its use in large-scale early screening. Photoplethysmography (PPG) has emerged as a promising screening modality due to its widespread availability in wearable devices and its ability to reflect underlying hemodynamic dynamics. However, the extreme scarcity of gold-standard labeled PPG data severely constrains the effectiveness of data-driven approaches. To address this challenge, we propose and validate a new paradigm, Physiology-Guided Self-Supervised Learning (PG-SSL), aimed at unlocking the value of large-scale unlabeled PPG data for efficient screening of Aortic Stenosis (AS) and Aortic Regurgitation (AR). Using over 170,000 unlabeled PPG samples from the UK Biobank, we formalize clinical knowledge into a set of PPG morphological phenotypes and construct a pulse pattern recognition proxy task for self-supervised pre-training. A dual-branch, gated-fusion architecture is then employed for efficient fine-tuning on a small labeled subset. The proposed PG-SSL framework achieves AUCs of 0.765 and 0.776 for AS and AR screening, respectively, significantly outperforming supervised baselines trained on limited labeled data. Multivariable analysis further validates the model output as an independent digital biomarker with sustained prognostic value after adjustment for standard clinical risk factors. This study demonstrates that PG-SSL provides an effective, domain knowledge-driven solution to label scarcity in medical artificial intelligence and shows strong potential for enabling low-cost, large-scale early screening of aortic valve disease.
SIGMA-PPG: Statistical-prior Informed Generative Masking Architecture for PPG Foundation Model
Jan 28, 2026Current foundation model for photoplethysmography (PPG) signals is challenged by the intrinsic redundancy and noise of the signal. Standard masked modeling often yields trivial solutions while contrastive methods lack morphological precision. To address these limitations, we propose a Statistical-prior Informed Generative Masking Architecture (SIGMA-PPG), a generative foundation model featuring a Prior-Guided Adversarial Masking mechanism, where a reinforcement learning-driven teacher leverages statistical priors to create challenging learning paths that prevent overfitting to noise. We also incorporate a semantic consistency constraint via vector quantization to ensure that physiologically identical waveforms (even those altered by recording artifacts or minor perturbations) map to shared indices. This enhances codebook semantic density and eliminates redundant feature structures. Pre-trained on over 120,000 hours of data, SIGMA-PPG achieves superior average performance compared to five state-of-the-art baselines across 12 diverse downstream tasks. The code is available at https://github.com/ZonghengGuo/SigmaPPG.
Facial Spatiotemporal Graphs: Leveraging the 3D Facial Surface for Remote Physiological Measurement
Jan 20, 2026Facial remote photoplethysmography (rPPG) methods estimate physiological signals by modeling subtle color changes on the 3D facial surface over time. However, existing methods fail to explicitly align their receptive fields with the 3D facial surface-the spatial support of the rPPG signal. To address this, we propose the Facial Spatiotemporal Graph (STGraph), a novel representation that encodes facial color and structure using 3D facial mesh sequences-enabling surface-aligned spatiotemporal processing. We introduce MeshPhys, a lightweight spatiotemporal graph convolutional network that operates on the STGraph to estimate physiological signals. Across four benchmark datasets, MeshPhys achieves state-of-the-art or competitive performance in both intra- and cross-dataset settings. Ablation studies show that constraining the model's receptive field to the facial surface acts as a strong structural prior, and that surface-aligned, 3D-aware node features are critical for robustly encoding facial surface color. Together, the STGraph and MeshPhys constitute a novel, principled modeling paradigm for facial rPPG, enabling robust, interpretable, and generalizable estimation. Code is available at https://samcantrill.github.io/facial-stgraph-rppg/ .
Wavelet-Driven Masked Multiscale Reconstruction for PPG Foundation Models
Jan 18, 2026Wearable foundation models have the potential to transform digital health by learning transferable representations from large-scale biosignals collected in everyday settings. While recent progress has been made in large-scale pretraining, most approaches overlook the spectral structure of photoplethysmography (PPG) signals, wherein physiological rhythms unfold across multiple frequency bands. Motivated by the insight that many downstream health-related tasks depend on multi-resolution features spanning fine-grained waveform morphology to global rhythmic dynamics, we introduce Masked Multiscale Reconstruction (MMR) for PPG representation learning - a self-supervised pretraining framework that explicitly learns from hierarchical time-frequency scales of PPG data. The pretraining task is designed to reconstruct randomly masked out coefficients obtained from a wavelet-based multiresolution decomposition of PPG signals, forcing the transformer encoder to integrate information across temporal and spectral scales. We pretrain our model with MMR using ~17 million unlabeled 10-second PPG segments from ~32,000 smartwatch users. On 17 of 19 diverse health-related tasks, MMR trained on large-scale wearable PPG data improves over or matches state-of-the-art open-source PPG foundation models, time-series foundation models, and other self-supervised baselines. Extensive analysis of our learned embeddings and systematic ablations underscores the value of wavelet-based representations, showing that they capture robust and physiologically-grounded features. Together, these results highlight the potential of MMR as a step toward generalizable PPG foundation models.
EvoMorph: Counterfactual Explanations for Continuous Time-Series Extrinsic Regression Applied to Photoplethysmography
Jan 15, 2026Wearable devices enable continuous, population-scale monitoring of physiological signals, such as photoplethysmography (PPG), creating new opportunities for data-driven clinical assessment. Time-series extrinsic regression (TSER) models increasingly leverage PPG signals to estimate clinically relevant outcomes, including heart rate, respiratory rate, and oxygen saturation. For clinical reasoning and trust, however, single point estimates alone are insufficient: clinicians must also understand whether predictions are stable under physiologically plausible variations and to what extent realistic, attainable changes in physiological signals would meaningfully alter a model's prediction. Counterfactual explanations (CFE) address these "what-if" questions, yet existing time series CFE generation methods are largely restricted to classification, overlook waveform morphology, and often produce physiologically implausible signals, limiting their applicability to continuous biomedical time series. To address these limitations, we introduce EvoMorph, a multi-objective evolutionary framework for generating physiologically plausible and diverse CFE for TSER applications. EvoMorph optimizes morphology-aware objectives defined on interpretable signal descriptors and applies transformations to preserve the waveform structure. We evaluated EvoMorph on three PPG datasets (heart rate, respiratory rate, and oxygen saturation) against a nearest-unlike-neighbor baseline. In addition, in a case study, we evaluated EvoMorph as a tool for uncertainty quantification by relating counterfactual sensitivity to bootstrap-ensemble uncertainty and data-density measures. Overall, EvoMorph enables the generation of physiologically-aware counterfactuals for continuous biomedical signals and supports uncertainty-aware interpretability, advancing trustworthy model analysis for clinical time-series applications.
ToTMNet: FFT-Accelerated Toeplitz Temporal Mixing Network for Lightweight Remote Photoplethysmography
Jan 07, 2026Remote photoplethysmography (rPPG) estimates a blood volume pulse (BVP) waveform from facial videos captured by commodity cameras. Although recent deep models improve robustness compared to classical signal-processing approaches, many methods increase computational cost and parameter count, and attention-based temporal modeling introduces quadratic scaling with respect to the temporal length. This paper proposes ToTMNet, a lightweight rPPG architecture that replaces temporal attention with an FFT-accelerated Toeplitz temporal mixing layer. The Toeplitz operator provides full-sequence temporal receptive field using a linear number of parameters in the clip length and can be applied in near-linear time using circulant embedding and FFT-based convolution. ToTMNet integrates the global Toeplitz temporal operator into a compact gated temporal mixer that combines a local depthwise temporal convolution branch with gated global Toeplitz mixing, enabling efficient long-range temporal filtering while only having 63k parameters. Experiments on two datasets, UBFC-rPPG (real videos) and SCAMPS (synthetic videos), show that ToTMNet achieves strong heart-rate estimation accuracy with a compact design. On UBFC-rPPG intra-dataset evaluation, ToTMNet reaches 1.055 bpm MAE with Pearson correlation 0.996. In a synthetic-to-real setting (SCAMPS to UBFC-rPPG), ToTMNet reaches 1.582 bpm MAE with Pearson correlation 0.994. Ablation results confirm that the gating mechanism is important for effectively using global Toeplitz mixing, especially under domain shift. The main limitation of this preprint study is the use of only two datasets; nevertheless, the results indicate that Toeplitz-structured temporal mixing is a practical and efficient alternative to attention for rPPG.
KDPhys: An Attention Guided 3D to 2D Knowledge Distillation for Real-time Video-Based Physiological Measurement
Jan 02, 2026Camera-based physiological monitoring, such as remote photoplethysmography (rPPG), captures subtle variations in skin optical properties caused by pulsatile blood volume changes using standard digital camera sensors. The demand for real-time, non-contact physiological measurement has increased significantly, particularly during the SARS-CoV-2 pandemic, to support telehealth and remote health monitoring applications. In this work, we propose an attention-based knowledge distillation (KD) framework, termed KDPhys, for extracting rPPG signals from facial video sequences. The proposed method distills global temporal representations from a 3D convolutional neural network (CNN) teacher model to a lightweight 2D CNN student model through effective 3D-to-2D feature distillation. To the best of our knowledge, this is the first application of knowledge distillation in the rPPG domain. Furthermore, we introduce a Distortion Loss incorporating Shape and Time (DILATE), which jointly accounts for both morphological and temporal characteristics of rPPG signals. Extensive qualitative and quantitative evaluations are conducted on three benchmark datasets. The proposed model achieves a significant reduction in computational complexity, using only half the parameters of existing methods while operating 56.67% faster. With just 0.23M parameters, it achieves an 18.15% reduction in Mean Absolute Error (MAE) compared to state-of-the-art approaches, attaining an average MAE of 1.78 bpm across all datasets. Additional experiments under diverse environmental conditions and activity scenarios further demonstrate the robustness and adaptability of the proposed approach.
* This paper has been published in Biomedical Signal Processing and Control
Quality-Aware Framework for Video-Derived Respiratory Signals
Dec 16, 2025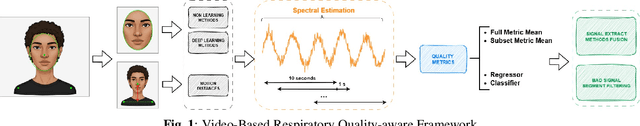
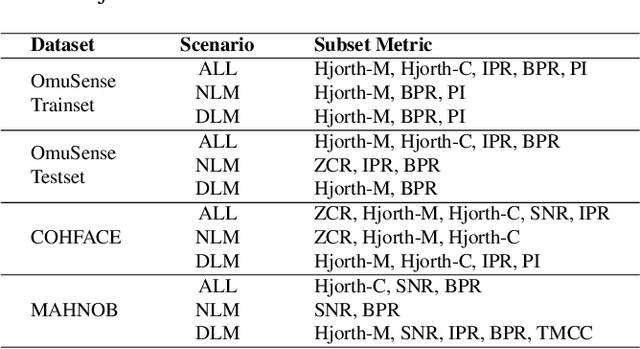
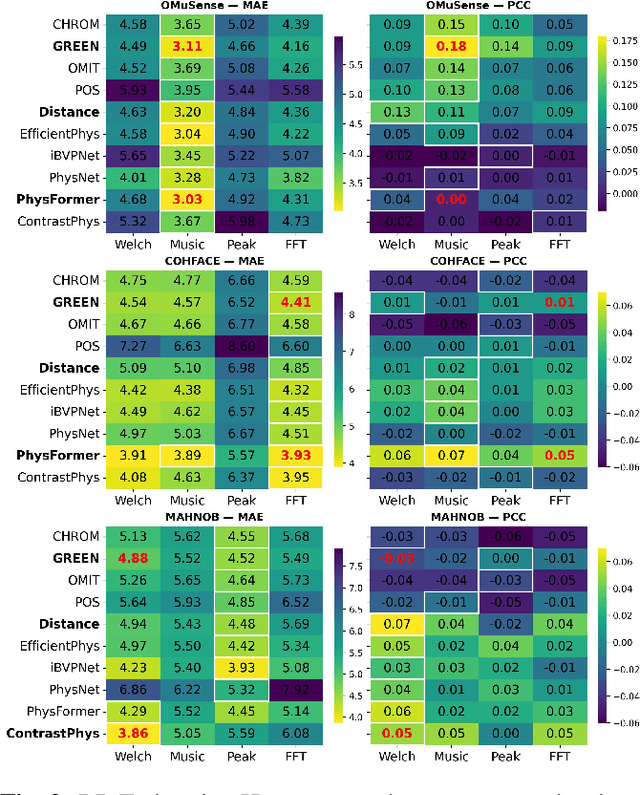
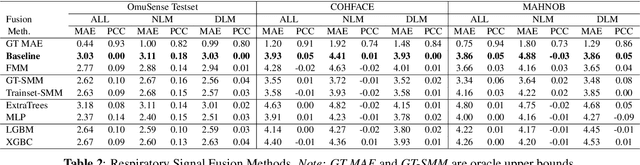
Video-based respiratory rate (RR) estimation is often unreliable due to inconsistent signal quality across extraction methods. We present a predictive, quality-aware framework that integrates heterogeneous signal sources with dynamic assessment of reliability. Ten signals are extracted from facial remote photoplethysmography (rPPG), upper-body motion, and deep learning pipelines, and analyzed using four spectral estimators: Welch's method, Multiple Signal Classification (MUSIC), Fast Fourier Transform (FFT), and peak detection. Segment-level quality indices are then used to train machine learning models that predict accuracy or select the most reliable signal. This enables adaptive signal fusion and quality-based segment filtering. Experiments on three public datasets (OMuSense-23, COHFACE, MAHNOB-HCI) show that the proposed framework achieves lower RR estimation errors than individual methods in most cases, with performance gains depending on dataset characteristics. These findings highlight the potential of quality-driven predictive modeling to deliver scalable and generalizable video-based respiratory monitoring solutions.
PMB-NN: Physiology-Centred Hybrid AI for Personalized Hemodynamic Monitoring from Photoplethysmography
Dec 11, 2025Continuous monitoring of blood pressure (BP) and hemodynamic parameters such as peripheral resistance (R) and arterial compliance (C) are critical for early vascular dysfunction detection. While photoplethysmography (PPG) wearables has gained popularity, existing data-driven methods for BP estimation lack interpretability. We advanced our previously proposed physiology-centered hybrid AI method-Physiological Model-Based Neural Network (PMB-NN)-in blood pressure estimation, that unifies deep learning with a 2-element Windkessel based model parameterized by R and C acting as physics constraints. The PMB-NN model was trained in a subject-specific manner using PPG-derived timing features, while demographic information was used to infer an intermediate variable: cardiac output. We validated the model on 10 healthy adults performing static and cycling activities across two days for model's day-to-day robustness, benchmarked against deep learning (DL) models (FCNN, CNN-LSTM, Transformer) and standalone Windkessel based physiological model (PM). Validation was conducted on three perspectives: accuracy, interpretability and plausibility. PMB-NN achieved systolic BP accuracy (MAE: 7.2 mmHg) comparable to DL benchmarks, diastolic performance (MAE: 3.9 mmHg) lower than DL models. However, PMB-NN exhibited higher physiological plausibility than both DL baselines and PM, suggesting that the hybrid architecture unifies and enhances the respective merits of physiological principles and data-driven techniques. Beyond BP, PMB-NN identified R (ME: 0.15 mmHg$\cdot$s/ml) and C (ME: -0.35 ml/mmHg) during training with accuracy similar to PM, demonstrating that the embedded physiological constraints confer interpretability to the hybrid AI framework. These results position PMB-NN as a balanced, physiologically grounded alternative to purely data-driven approaches for daily hemodynamic monitoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge