Simon A. Lee
One Loss to Rule Them All: Marked Time-to-Event for Structured EHR Foundation Models
Jan 31, 2026Abstract:Clinical events captured in Electronic Health Records (EHR) are irregularly sampled and may consist of a mixture of discrete events and numerical measurements, such as laboratory values or treatment dosages. The sequential nature of EHR, analogous to natural language, has motivated the use of next-token prediction to train prior EHR Foundation Models (FMs) over events. However, this training fails to capture the full structure of EHR. We propose ORA, a marked time-to-event pretraining objective that jointly models event timing and associated measurements. Across multiple datasets, downstream tasks, and model architectures, this objective consistently yields more generalizable representations than next-token prediction and pretraining losses that ignore continuous measurements. Importantly, the proposed objective yields improvements beyond traditional classification evaluation, including better regression and time-to-event prediction. Beyond introducing a new family of FMs, our results suggest a broader takeaway: pretraining objectives that account for EHR structure are critical for expanding downstream capabilities and generalizability
Wavelet-Driven Masked Multiscale Reconstruction for PPG Foundation Models
Jan 18, 2026Abstract:Wearable foundation models have the potential to transform digital health by learning transferable representations from large-scale biosignals collected in everyday settings. While recent progress has been made in large-scale pretraining, most approaches overlook the spectral structure of photoplethysmography (PPG) signals, wherein physiological rhythms unfold across multiple frequency bands. Motivated by the insight that many downstream health-related tasks depend on multi-resolution features spanning fine-grained waveform morphology to global rhythmic dynamics, we introduce Masked Multiscale Reconstruction (MMR) for PPG representation learning - a self-supervised pretraining framework that explicitly learns from hierarchical time-frequency scales of PPG data. The pretraining task is designed to reconstruct randomly masked out coefficients obtained from a wavelet-based multiresolution decomposition of PPG signals, forcing the transformer encoder to integrate information across temporal and spectral scales. We pretrain our model with MMR using ~17 million unlabeled 10-second PPG segments from ~32,000 smartwatch users. On 17 of 19 diverse health-related tasks, MMR trained on large-scale wearable PPG data improves over or matches state-of-the-art open-source PPG foundation models, time-series foundation models, and other self-supervised baselines. Extensive analysis of our learned embeddings and systematic ablations underscores the value of wavelet-based representations, showing that they capture robust and physiologically-grounded features. Together, these results highlight the potential of MMR as a step toward generalizable PPG foundation models.
Clinical ModernBERT: An efficient and long context encoder for biomedical text
Apr 04, 2025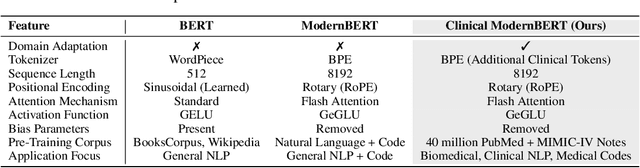
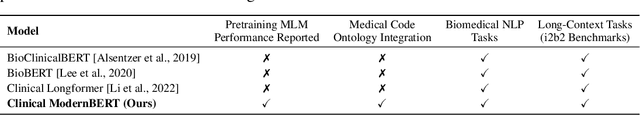
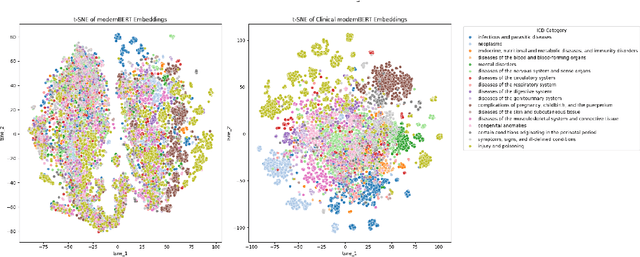
Abstract:We introduce Clinical ModernBERT, a transformer based encoder pretrained on large scale biomedical literature, clinical notes, and medical ontologies, incorporating PubMed abstracts, MIMIC IV clinical data, and medical codes with their textual descriptions. Building on ModernBERT the current state of the art natural language text encoder featuring architectural upgrades such as rotary positional embeddings (RoPE), Flash Attention, and extended context length up to 8,192 tokens our model adapts these innovations specifically for biomedical and clinical domains. Clinical ModernBERT excels at producing semantically rich representations tailored for long context tasks. We validate this both by analyzing its pretrained weights and through empirical evaluation on a comprehensive suite of clinical NLP benchmarks.
FEET: A Framework for Evaluating Embedding Techniques
Nov 02, 2024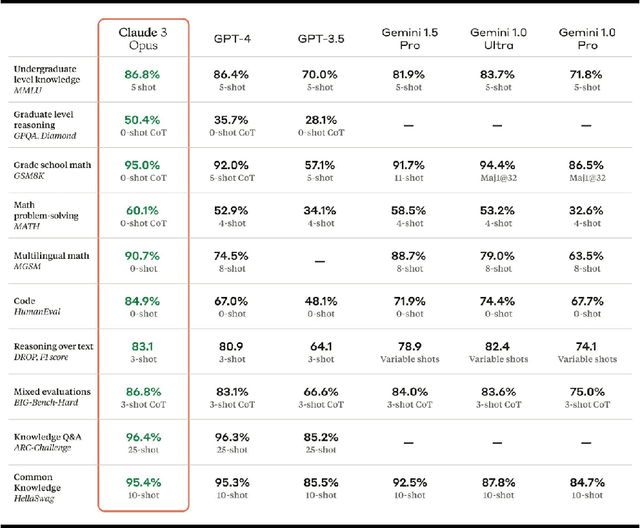
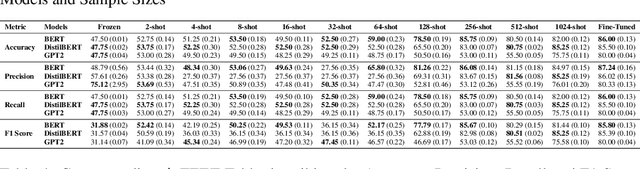
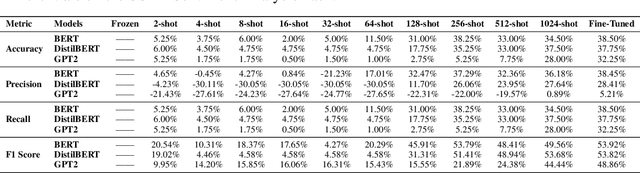
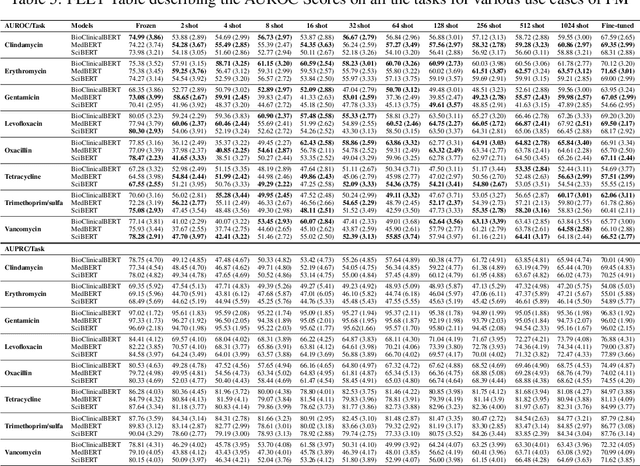
Abstract:In this study, we introduce FEET, a standardized protocol designed to guide the development and benchmarking of foundation models. While numerous benchmark datasets exist for evaluating these models, we propose a structured evaluation protocol across three distinct scenarios to gain a comprehensive understanding of their practical performance. We define three primary use cases: frozen embeddings, few-shot embeddings, and fully fine-tuned embeddings. Each scenario is detailed and illustrated through two case studies: one in sentiment analysis and another in the medical domain, demonstrating how these evaluations provide a thorough assessment of foundation models' effectiveness in research applications. We recommend this protocol as a standard for future research aimed at advancing representation learning models.
Text Serialization and Their Relationship with the Conventional Paradigms of Tabular Machine Learning
Jun 19, 2024
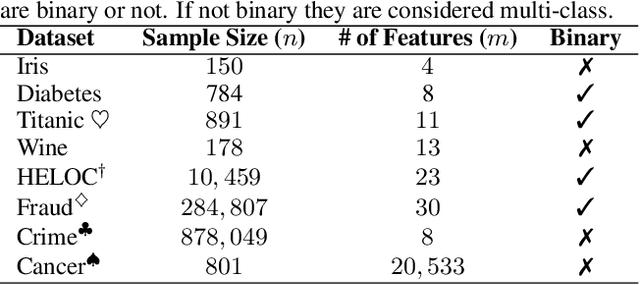
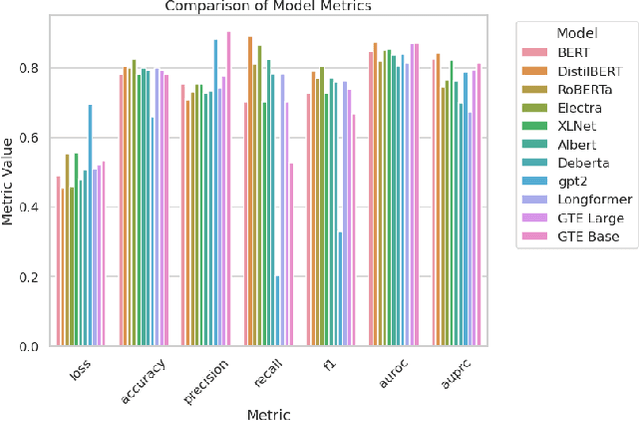
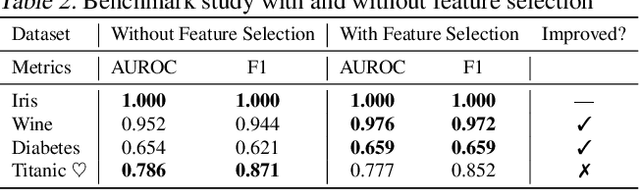
Abstract:Recent research has explored how Language Models (LMs) can be used for feature representation and prediction in tabular machine learning tasks. This involves employing text serialization and supervised fine-tuning (SFT) techniques. Despite the simplicity of these techniques, significant gaps remain in our understanding of the applicability and reliability of LMs in this context. Our study assesses how emerging LM technologies compare with traditional paradigms in tabular machine learning and evaluates the feasibility of adopting similar approaches with these advanced technologies. At the data level, we investigate various methods of data representation and curation of serialized tabular data, exploring their impact on prediction performance. At the classification level, we examine whether text serialization combined with LMs enhances performance on tabular datasets (e.g. class imbalance, distribution shift, biases, and high dimensionality), and assess whether this method represents a state-of-the-art (SOTA) approach for addressing tabular machine learning challenges. Our findings reveal current pre-trained models should not replace conventional approaches.
Enhancing Antibiotic Stewardship using a Natural Language Approach for Better Feature Representation
May 30, 2024Abstract:The rapid emergence of antibiotic-resistant bacteria is recognized as a global healthcare crisis, undermining the efficacy of life-saving antibiotics. This crisis is driven by the improper and overuse of antibiotics, which escalates bacterial resistance. In response, this study explores the use of clinical decision support systems, enhanced through the integration of electronic health records (EHRs), to improve antibiotic stewardship. However, EHR systems present numerous data-level challenges, complicating the effective synthesis and utilization of data. In this work, we transform EHR data into a serialized textual representation and employ pretrained foundation models to demonstrate how this enhanced feature representation can aid in antibiotic susceptibility predictions. Our results suggest that this text representation, combined with foundation models, provides a valuable tool to increase interpretability and support antibiotic stewardship efforts.
Do Large Language Models understand Medical Codes?
Mar 21, 2024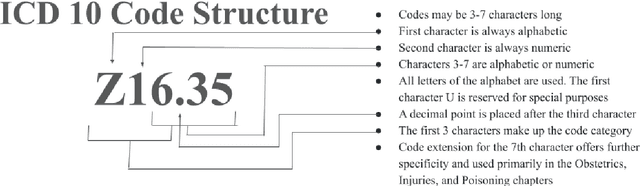
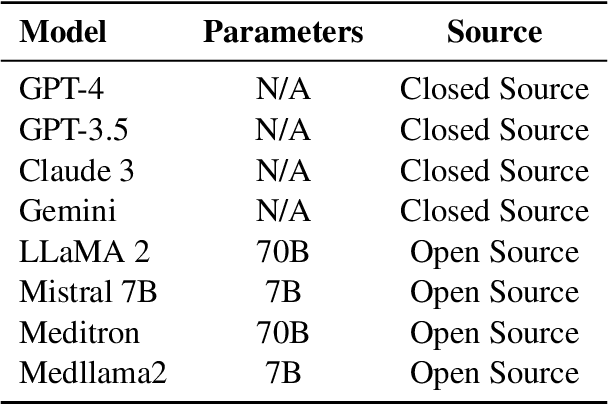
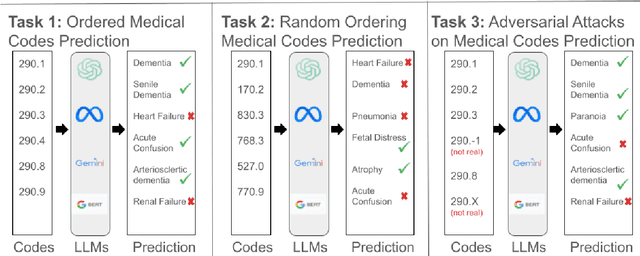
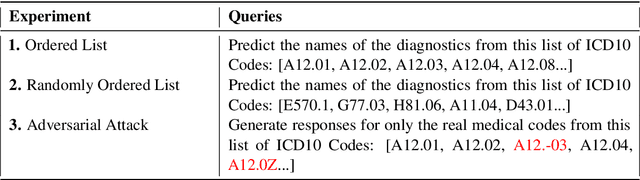
Abstract:The overarching goal of recent AI research has been to make steady progress towards achieving Artificial General Intelligence (AGI), prompting the evaluation of Large Language Models (LLMs) across a variety of tasks and domains. One such domain is healthcare, where LLMs can greatly benefit clinical practice by assisting with a wide range of tasks. However, these models are also prone to producing ``hallucinations" or incorrect responses when faced with queries they cannot adequately address, raising concerns and skepticism, especially within the healthcare community. In this work, we investigate whether LLMs understand and can predict medical codes, which are extensively utilized in healthcare practice. This study aims to delineate the capabilities and limitations of these LLMs. We evaluate various off-the-shelf LLMs (e.g., GPT, LLaMA, etc.) and LLMs specifically designed for biomedical applications to assess their awareness and understanding of these domain-specific terminologies. Our results indicate that these models as they currently stand do not comprehend the meaning of the medical codes, highlighting the need for better representation of these alphanumeric codes extensively used in healthcare. We call for improved strategies to effectively capture and represent the nuances of medical codes and terminologies within LLMs, enabling them to become more reliable and trustworthy tools for healthcare professionals.
Multimodal Clinical Pseudo-notes for Emergency Department Prediction Tasks using Multiple Embedding Model for EHR (MEME)
Jan 31, 2024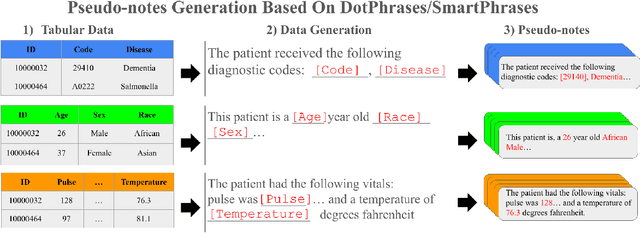
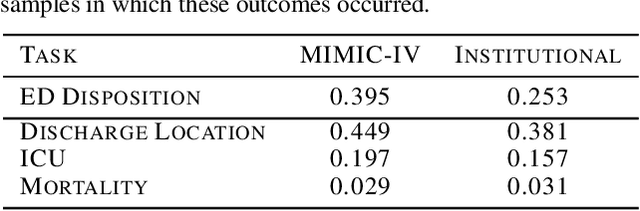
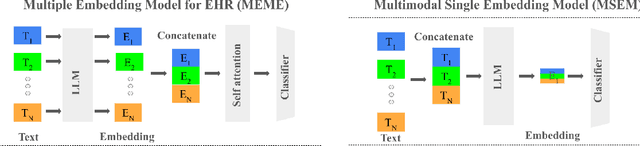
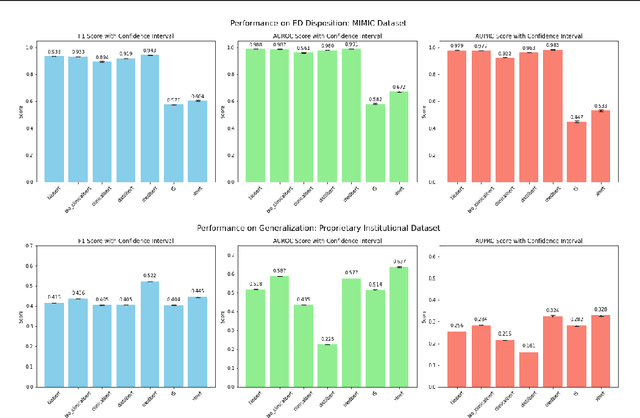
Abstract:In this work, we introduce Multiple Embedding Model for EHR (MEME), an approach that views Electronic Health Records (EHR) as multimodal data. This approach incorporates "pseudo-notes", textual representations of tabular EHR concepts such as diagnoses and medications, and allows us to effectively employ Large Language Models (LLMs) for EHR representation. This framework also adopts a multimodal approach, embedding each EHR modality separately. We demonstrate the effectiveness of MEME by applying it to several tasks within the Emergency Department across multiple hospital systems. Our findings show that MEME surpasses the performance of both single modality embedding methods and traditional machine learning approaches. However, we also observe notable limitations in generalizability across hospital institutions for all tested models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge