Mohanasankar Sivaprakasam
CytoCLIP: Learning Cytoarchitectural Characteristics in Developing Human Brain Using Contrastive Language Image Pre-Training
Jan 18, 2026Abstract:The functions of different regions of the human brain are closely linked to their distinct cytoarchitecture, which is defined by the spatial arrangement and morphology of the cells. Identifying brain regions by their cytoarchitecture enables various scientific analyses of the brain. However, delineating these areas manually in brain histological sections is time-consuming and requires specialized knowledge. An automated approach is necessary to minimize the effort needed from human experts. To address this, we propose CytoCLIP, a suite of vision-language models derived from pre-trained Contrastive Language-Image Pre-Training (CLIP) frameworks to learn joint visual-text representations of brain cytoarchitecture. CytoCLIP comprises two model variants: one is trained using low-resolution whole-region images to understand the overall cytoarchitectural pattern of an area, and the other is trained on high-resolution image tiles for detailed cellular-level representation. The training dataset is created from NISSL-stained histological sections of developing fetal brains of different gestational weeks. It includes 86 distinct regions for low-resolution images and 384 brain regions for high-resolution tiles. We evaluate the model's understanding of the cytoarchitecture and generalization ability using region classification and cross-modal retrieval tasks. Multiple experiments are performed under various data setups, including data from samples of different ages and sectioning planes. Experimental results demonstrate that CytoCLIP outperforms existing methods. It achieves an F1 score of 0.87 for whole-region classification and 0.91 for high-resolution image tile classification.
KDPhys: An Attention Guided 3D to 2D Knowledge Distillation for Real-time Video-Based Physiological Measurement
Jan 02, 2026Abstract:Camera-based physiological monitoring, such as remote photoplethysmography (rPPG), captures subtle variations in skin optical properties caused by pulsatile blood volume changes using standard digital camera sensors. The demand for real-time, non-contact physiological measurement has increased significantly, particularly during the SARS-CoV-2 pandemic, to support telehealth and remote health monitoring applications. In this work, we propose an attention-based knowledge distillation (KD) framework, termed KDPhys, for extracting rPPG signals from facial video sequences. The proposed method distills global temporal representations from a 3D convolutional neural network (CNN) teacher model to a lightweight 2D CNN student model through effective 3D-to-2D feature distillation. To the best of our knowledge, this is the first application of knowledge distillation in the rPPG domain. Furthermore, we introduce a Distortion Loss incorporating Shape and Time (DILATE), which jointly accounts for both morphological and temporal characteristics of rPPG signals. Extensive qualitative and quantitative evaluations are conducted on three benchmark datasets. The proposed model achieves a significant reduction in computational complexity, using only half the parameters of existing methods while operating 56.67% faster. With just 0.23M parameters, it achieves an 18.15% reduction in Mean Absolute Error (MAE) compared to state-of-the-art approaches, attaining an average MAE of 1.78 bpm across all datasets. Additional experiments under diverse environmental conditions and activity scenarios further demonstrate the robustness and adaptability of the proposed approach.
* This paper has been published in Biomedical Signal Processing and Control
AD-DAE: Unsupervised Modeling of Longitudinal Alzheimer's Disease Progression with Diffusion Auto-Encoder
Nov 08, 2025Abstract:Generative modeling frameworks have emerged as an effective approach to capture high-dimensional image distributions from large datasets without requiring domain-specific knowledge, a capability essential for longitudinal disease progression modeling. Recent generative modeling approaches have attempted to capture progression by mapping images into a latent representational space and then controlling and guiding the representations to generate follow-up images from a baseline image. However, existing approaches impose constraints on distribution learning, leading to latent spaces with limited controllability to generate follow-up images without explicit supervision from subject-specific longitudinal images. In order to enable controlled movements in the latent representational space and generate progression images from a baseline image in an unsupervised manner, we introduce a conditionable Diffusion Auto-encoder framework. The explicit encoding mechanism of image-diffusion auto-encoders forms a compact latent space capturing high-level semantics, providing means to disentangle information relevant for progression. Our approach leverages this latent space to condition and apply controlled shifts to baseline representations for generating follow-up. Controllability is induced by restricting these shifts to a subspace, thereby isolating progression-related factors from subject identity-preserving components. The shifts are implicitly guided by correlating with progression attributes, without requiring subject-specific longitudinal supervision. We validate the generations through image quality metrics, volumetric progression analysis, and downstream classification in Alzheimer's disease datasets from two different sources and disease categories. This demonstrates the effectiveness of our approach for Alzheimer's progression modeling and longitudinal image generation.
PosDiffAE: Position-aware Diffusion Auto-encoder For High-Resolution Brain Tissue Classification Incorporating Artifact Restoration
Jul 03, 2025Abstract:Denoising diffusion models produce high-fidelity image samples by capturing the image distribution in a progressive manner while initializing with a simple distribution and compounding the distribution complexity. Although these models have unlocked new applicabilities, the sampling mechanism of diffusion does not offer means to extract image-specific semantic representation, which is inherently provided by auto-encoders. The encoding component of auto-encoders enables mapping between a specific image and its latent space, thereby offering explicit means of enforcing structures in the latent space. By integrating an encoder with the diffusion model, we establish an auto-encoding formulation, which learns image-specific representations and offers means to organize the latent space. In this work, First, we devise a mechanism to structure the latent space of a diffusion auto-encoding model, towards recognizing region-specific cellular patterns in brain images. We enforce the representations to regress positional information of the patches from high-resolution images. This creates a conducive latent space for differentiating tissue types of the brain. Second, we devise an unsupervised tear artifact restoration technique based on neighborhood awareness, utilizing latent representations and the constrained generation capability of diffusion models during inference. Third, through representational guidance and leveraging the inference time steerable noising and denoising capability of diffusion, we devise an unsupervised JPEG artifact restoration technique.
Position Paper: Building Trust in Synthetic Data for Clinical AI
Feb 04, 2025Abstract:Deep generative models and synthetic medical data have shown significant promise in addressing key challenges in healthcare, such as privacy concerns, data bias, and the scarcity of realistic datasets. While research in this area has grown rapidly and demonstrated substantial theoretical potential, its practical adoption in clinical settings remains limited. Despite the benefits synthetic data offers, questions surrounding its reliability and credibility persist, leading to a lack of trust among clinicians. This position paper argues that fostering trust in synthetic medical data is crucial for its clinical adoption. It aims to spark a discussion on the viability of synthetic medical data in clinical practice, particularly in the context of current advancements in AI. We present empirical evidence from brain tumor segmentation to demonstrate that the quality, diversity, and proportion of synthetic data directly impact trust in clinical AI models. Our findings provide insights to improve the deployment and acceptance of synthetic data-driven AI systems in real-world clinical workflows.
AAD-DCE: An Aggregated Multimodal Attention Mechanism for Early and Late Dynamic Contrast Enhanced Prostate MRI Synthesis
Feb 04, 2025Abstract:Dynamic Contrast-Enhanced Magnetic Resonance Imaging (DCE-MRI) is a medical imaging technique that plays a crucial role in the detailed visualization and identification of tissue perfusion in abnormal lesions and radiological suggestions for biopsy. However, DCE-MRI involves the administration of a Gadolinium based (Gad) contrast agent, which is associated with a risk of toxicity in the body. Previous deep learning approaches that synthesize DCE-MR images employ unimodal non-contrast or low-dose contrast MRI images lacking focus on the local perfusion information within the anatomy of interest. We propose AAD-DCE, a generative adversarial network (GAN) with an aggregated attention discriminator module consisting of global and local discriminators. The discriminators provide a spatial embedded attention map to drive the generator to synthesize early and late response DCE-MRI images. Our method employs multimodal inputs - T2 weighted (T2W), Apparent Diffusion Coefficient (ADC), and T1 pre-contrast for image synthesis. Extensive comparative and ablation studies on the ProstateX dataset show that our model (i) is agnostic to various generator benchmarks and (ii) outperforms other DCE-MRI synthesis approaches with improvement margins of +0.64 dB PSNR, +0.0518 SSIM, -0.015 MAE for early response and +0.1 dB PSNR, +0.0424 SSIM, -0.021 MAE for late response, and (ii) emphasize the importance of attention ensembling. Our code is available at https://github.com/bhartidivya/AAD-DCE.
Knowledge Models for Cancer Clinical Practice Guidelines : Construction, Management and Usage in Question Answering
Jul 23, 2024Abstract:An automated knowledge modeling algorithm for Cancer Clinical Practice Guidelines (CPGs) extracts the knowledge contained in the CPG documents and transforms it into a programmatically interactable, easy-to-update structured model with minimal human intervention. The existing automated algorithms have minimal scope and cannot handle the varying complexity of the knowledge content in the CPGs for different cancer types. This work proposes an improved automated knowledge modeling algorithm to create knowledge models from the National Comprehensive Cancer Network (NCCN) CPGs in Oncology for different cancer types. The proposed algorithm has been evaluated with NCCN CPGs for four different cancer types. We also proposed an algorithm to compare the knowledge models for different versions of a guideline to discover the specific changes introduced in the treatment protocol of a new version. We created a question-answering (Q&A) framework with the guideline knowledge models as the augmented knowledge base to study our ability to query the knowledge models. We compiled a set of 32 question-answer pairs derived from two reliable data sources for the treatment of Non-Small Cell Lung Cancer (NSCLC) to evaluate the Q&A framework. The framework was evaluated against the question-answer pairs from one data source, and it can generate the answers with 54.5% accuracy from the treatment algorithm and 81.8% accuracy from the discussion part of the NCCN NSCLC guideline knowledge model.
A Hybrid-Layered System for Image-Guided Navigation and Robot Assisted Spine Surgery
Jun 07, 2024Abstract:In response to the growing demand for precise and affordable solutions for Image-Guided Spine Surgery (IGSS), this paper presents a comprehensive development of a Robot-Assisted and Navigation-Guided IGSS System. The endeavor involves integrating cutting-edge technologies to attain the required surgical precision and limit user radiation exposure, thereby addressing the limitations of manual surgical methods. We propose an IGSS workflow and system architecture employing a hybrid-layered approach, combining modular and integrated system architectures in distinctive layers to develop an affordable system for seamless integration, scalability, and reconfigurability. We developed and integrated the system and extensively tested it on phantoms and cadavers. The proposed system's accuracy using navigation guidance is 1.020 mm, and robot assistance is 1.11 mm on phantoms. Observing a similar performance in cadaveric validation where 84% of screw placements were grade A, 10% were grade B using navigation guidance, 90% were grade A, and 10% were grade B using robot assistance as per the Gertzbein-Robbins scale, proving its efficacy for an IGSS. The evaluated performance is adequate for an IGSS and at par with the existing systems in literature and those commercially available. The user radiation is lower than in the literature, given that the system requires only an average of 3 C-Arm images per pedicle screw placement and verification
* 6 Pages, 4 Figures, Published in IEEE SII Conference
Online Action Representation using Change Detection and Symbolic Programming
May 19, 2024Abstract:This paper addresses the critical need for online action representation, which is essential for various applications like rehabilitation, surveillance, etc. The task can be defined as representation of actions as soon as they happen in a streaming video without access to video frames in the future. Most of the existing methods use predefined window sizes for video segments, which is a restrictive assumption on the dynamics. The proposed method employs a change detection algorithm to automatically segment action sequences, which form meaningful sub-actions and subsequently fit symbolic generative motion programs to the clipped segments. We determine the start time and end time of segments using change detection followed by a piece-wise linear fit algorithm on joint angle and bone length sequences. Domain-specific symbolic primitives are fit to pose keypoint trajectories of those extracted segments in order to obtain a higher level semantic representation. Since this representation is part-based, it is complementary to the compositional nature of human actions, i.e., a complex activity can be broken down into elementary sub-actions. We show the effectiveness of this representation in the downstream task of class agnostic repetition detection. We propose a repetition counting algorithm based on consecutive similarity matching of primitives, which can do online repetition counting. We also compare the results with a similar but offline repetition counting algorithm. The results of the experiments demonstrate that, despite operating online, the proposed method performs better or on par with the existing method.
DCE-FORMER: A Transformer-based Model With Mutual Information And Frequency-based Loss Functions For Early And Late Response Prediction In Prostate DCE-MRI
Feb 03, 2024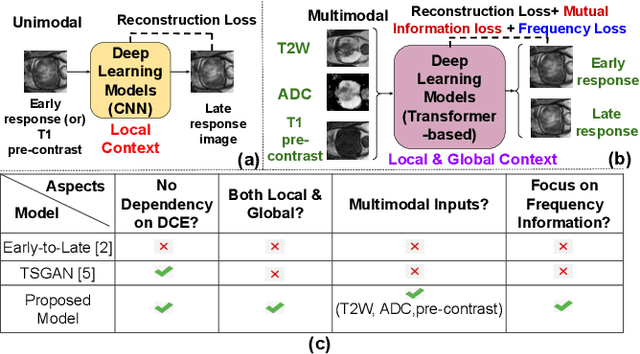
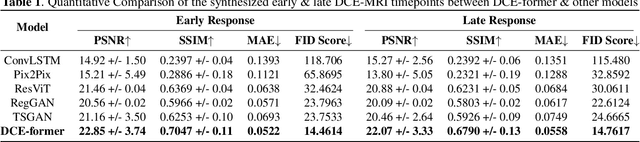
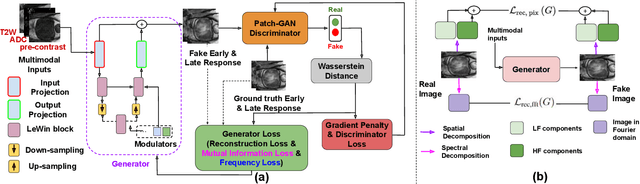
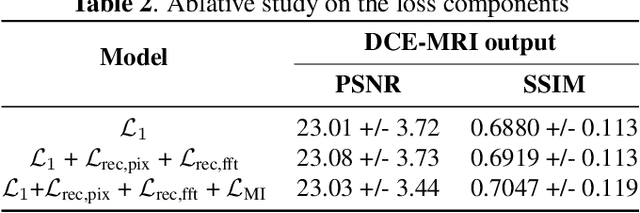
Abstract:Dynamic Contrast Enhanced Magnetic Resonance Imaging aids in the detection and assessment of tumor aggressiveness by using a Gadolinium-based contrast agent (GBCA). However, GBCA is known to have potential toxic effects. This risk can be avoided if we obtain DCE-MRI images without using GBCA. We propose, DCE-former, a transformer-based neural network to generate early and late response prostate DCE-MRI images from non-contrast multimodal inputs (T2 weighted, Apparent Diffusion Coefficient, and T1 pre-contrast MRI). Additionally, we introduce (i) a mutual information loss function to capture the complementary information about contrast uptake, and (ii) a frequency-based loss function in the pixel and Fourier space to learn local and global hyper-intensity patterns in DCE-MRI. Extensive experiments show that DCE-former outperforms other methods with improvement margins of +1.39 dB and +1.19 db in PSNR, +0.068 and +0.055 in SSIM, and -0.012 and -0.013 in Mean Absolute Error for early and late response DCE-MRI, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge