Mohammad Al Fahim
Geometric Learning-Based Transformer Network for Estimation of Segmentation Errors
Aug 10, 2023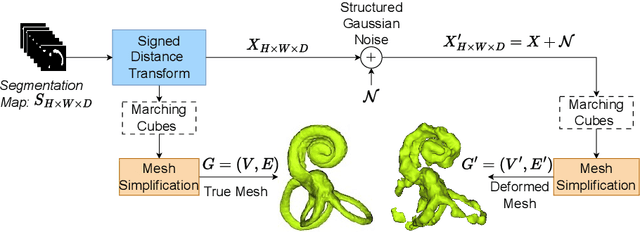


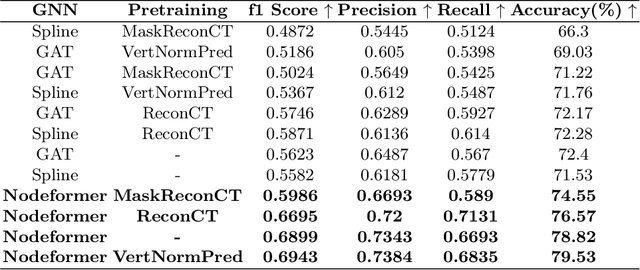
Abstract:Many segmentation networks have been proposed for 3D volumetric segmentation of tumors and organs at risk. Hospitals and clinical institutions seek to accelerate and minimize the efforts of specialists in image segmentation. Still, in case of errors generated by these networks, clinicians would have to manually edit the generated segmentation maps. Given a 3D volume and its putative segmentation map, we propose an approach to identify and measure erroneous regions in the segmentation map. Our method can estimate error at any point or node in a 3D mesh generated from a possibly erroneous volumetric segmentation map, serving as a Quality Assurance tool. We propose a graph neural network-based transformer based on the Nodeformer architecture to measure and classify the segmentation errors at any point. We have evaluated our network on a high-resolution micro-CT dataset of the human inner-ear bony labyrinth structure by simulating erroneous 3D segmentation maps. Our network incorporates a convolutional encoder to compute node-centric features from the input micro-CT data, the Nodeformer to learn the latent graph embeddings, and a Multi-Layer Perceptron (MLP) to compute and classify the node-wise errors. Our network achieves a mean absolute error of ~0.042 over other Graph Neural Networks (GNN) and an accuracy of 79.53% over other GNNs in estimating and classifying the node-wise errors, respectively. We also put forth vertex-normal prediction as a custom pretext task for pre-training the CNN encoder to improve the network's overall performance. Qualitative analysis shows the efficiency of our network in correctly classifying errors and reducing misclassifications.
HyperCoil-Recon: A Hypernetwork-based Adaptive Coil Configuration Task Switching Network for MRI Reconstruction
Aug 09, 2023

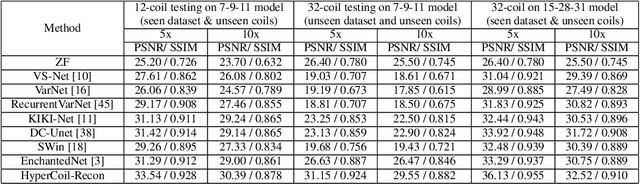

Abstract:Parallel imaging, a fast MRI technique, involves dynamic adjustments based on the configuration i.e. number, positioning, and sensitivity of the coils with respect to the anatomy under study. Conventional deep learning-based image reconstruction models have to be trained or fine-tuned for each configuration, posing a barrier to clinical translation, given the lack of computational resources and machine learning expertise for clinicians to train models at deployment. Joint training on diverse datasets learns a single weight set that might underfit to deviated configurations. We propose, HyperCoil-Recon, a hypernetwork-based coil configuration task-switching network for multi-coil MRI reconstruction that encodes varying configurations of the numbers of coils in a multi-tasking perspective, posing each configuration as a task. The hypernetworks infer and embed task-specific weights into the reconstruction network, 1) effectively utilizing the contextual knowledge of common and varying image features among the various fields-of-view of the coils, and 2) enabling generality to unseen configurations at test time. Experiments reveal that our approach 1) adapts on the fly to various unseen configurations up to 32 coils when trained on lower numbers (i.e. 7 to 11) of randomly varying coils, and to 120 deviated unseen configurations when trained on 18 configurations in a single model, 2) matches the performance of coil configuration-specific models, and 3) outperforms configuration-invariant models with improvement margins of around 1 dB / 0.03 and 0.3 dB / 0.02 in PSNR / SSIM for knee and brain data. Our code is available at https://github.com/sriprabhar/HyperCoil-Recon
SDLFormer: A Sparse and Dense Locality-enhanced Transformer for Accelerated MR Image Reconstruction
Aug 08, 2023



Abstract:Transformers have emerged as viable alternatives to convolutional neural networks owing to their ability to learn non-local region relationships in the spatial domain. The self-attention mechanism of the transformer enables transformers to capture long-range dependencies in the images, which might be desirable for accelerated MRI image reconstruction as the effect of undersampling is non-local in the image domain. Despite its computational efficiency, the window-based transformers suffer from restricted receptive fields as the dependencies are limited to within the scope of the image windows. We propose a window-based transformer network that integrates dilated attention mechanism and convolution for accelerated MRI image reconstruction. The proposed network consists of dilated and dense neighborhood attention transformers to enhance the distant neighborhood pixel relationship and introduce depth-wise convolutions within the transformer module to learn low-level translation invariant features for accelerated MRI image reconstruction. The proposed model is trained in a self-supervised manner. We perform extensive experiments for multi-coil MRI acceleration for coronal PD, coronal PDFS and axial T2 contrasts with 4x and 5x under-sampling in self-supervised learning based on k-space splitting. We compare our method against other reconstruction architectures and the parallel domain self-supervised learning baseline. Results show that the proposed model exhibits improvement margins of (i) around 1.40 dB in PSNR and around 0.028 in SSIM on average over other architectures (ii) around 1.44 dB in PSNR and around 0.029 in SSIM over parallel domain self-supervised learning. The code is available at https://github.com/rahul-gs-16/sdlformer.git
SFT-KD-Recon: Learning a Student-friendly Teacher for Knowledge Distillation in Magnetic Resonance Image Reconstruction
Apr 11, 2023



Abstract:Deep cascaded architectures for magnetic resonance imaging (MRI) acceleration have shown remarkable success in providing high-quality reconstruction. However, as the number of cascades increases, the improvements in reconstruction tend to become marginal, indicating possible excess model capacity. Knowledge distillation (KD) is an emerging technique to compress these models, in which a trained deep teacher network is used to distill knowledge to a smaller student network such that the student learns to mimic the behavior of the teacher. Most KD methods focus on effectively training the student with a pre-trained teacher unaware of the student model. We propose SFT-KD-Recon, a student-friendly teacher training approach along with the student as a prior step to KD to make the teacher aware of the structure and capacity of the student and enable aligning the representations of the teacher with the student. In SFT, the teacher is jointly trained with the unfolded branch configurations of the student blocks using three loss terms - teacher-reconstruction loss, student-reconstruction loss, and teacher-student imitation loss, followed by KD of the student. We perform extensive experiments for MRI acceleration in 4x and 5x under-sampling on the brain and cardiac datasets on five KD methods using the proposed approach as a prior step. We consider the DC-CNN architecture and setup teacher as D5C5 (141765 parameters), and student as D3C5 (49285 parameters), denoting a compression of 2.87:1. Results show that (i) our approach consistently improves the KD methods with improved reconstruction performance and image quality, and (ii) the student distilled using our approach is competitive with the teacher, with the performance gap reduced from 0.53 dB to 0.03 dB.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge