Pralaypati Ta
CytoCLIP: Learning Cytoarchitectural Characteristics in Developing Human Brain Using Contrastive Language Image Pre-Training
Jan 18, 2026Abstract:The functions of different regions of the human brain are closely linked to their distinct cytoarchitecture, which is defined by the spatial arrangement and morphology of the cells. Identifying brain regions by their cytoarchitecture enables various scientific analyses of the brain. However, delineating these areas manually in brain histological sections is time-consuming and requires specialized knowledge. An automated approach is necessary to minimize the effort needed from human experts. To address this, we propose CytoCLIP, a suite of vision-language models derived from pre-trained Contrastive Language-Image Pre-Training (CLIP) frameworks to learn joint visual-text representations of brain cytoarchitecture. CytoCLIP comprises two model variants: one is trained using low-resolution whole-region images to understand the overall cytoarchitectural pattern of an area, and the other is trained on high-resolution image tiles for detailed cellular-level representation. The training dataset is created from NISSL-stained histological sections of developing fetal brains of different gestational weeks. It includes 86 distinct regions for low-resolution images and 384 brain regions for high-resolution tiles. We evaluate the model's understanding of the cytoarchitecture and generalization ability using region classification and cross-modal retrieval tasks. Multiple experiments are performed under various data setups, including data from samples of different ages and sectioning planes. Experimental results demonstrate that CytoCLIP outperforms existing methods. It achieves an F1 score of 0.87 for whole-region classification and 0.91 for high-resolution image tile classification.
Knowledge Models for Cancer Clinical Practice Guidelines : Construction, Management and Usage in Question Answering
Jul 23, 2024Abstract:An automated knowledge modeling algorithm for Cancer Clinical Practice Guidelines (CPGs) extracts the knowledge contained in the CPG documents and transforms it into a programmatically interactable, easy-to-update structured model with minimal human intervention. The existing automated algorithms have minimal scope and cannot handle the varying complexity of the knowledge content in the CPGs for different cancer types. This work proposes an improved automated knowledge modeling algorithm to create knowledge models from the National Comprehensive Cancer Network (NCCN) CPGs in Oncology for different cancer types. The proposed algorithm has been evaluated with NCCN CPGs for four different cancer types. We also proposed an algorithm to compare the knowledge models for different versions of a guideline to discover the specific changes introduced in the treatment protocol of a new version. We created a question-answering (Q&A) framework with the guideline knowledge models as the augmented knowledge base to study our ability to query the knowledge models. We compiled a set of 32 question-answer pairs derived from two reliable data sources for the treatment of Non-Small Cell Lung Cancer (NSCLC) to evaluate the Q&A framework. The framework was evaluated against the question-answer pairs from one data source, and it can generate the answers with 54.5% accuracy from the treatment algorithm and 81.8% accuracy from the discussion part of the NCCN NSCLC guideline knowledge model.
Automated Knowledge Modeling for Cancer Clinical Practice Guidelines
Jul 15, 2023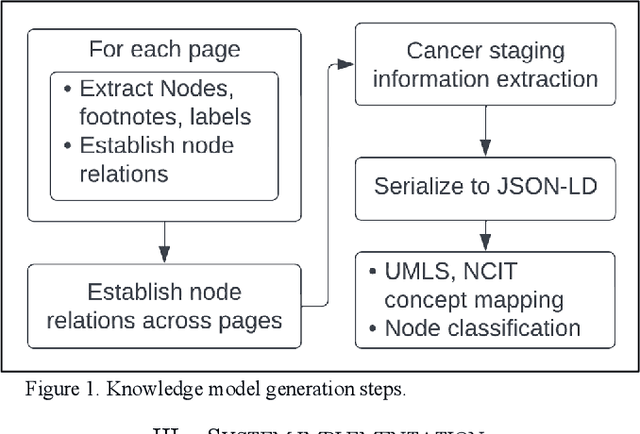
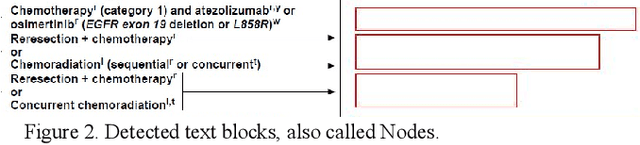
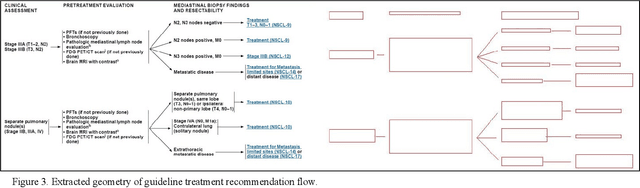
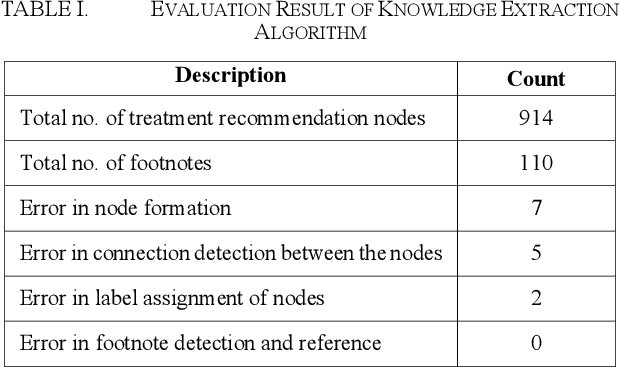
Abstract:Clinical Practice Guidelines (CPGs) for cancer diseases evolve rapidly due to new evidence generated by active research. Currently, CPGs are primarily published in a document format that is ill-suited for managing this developing knowledge. A knowledge model of the guidelines document suitable for programmatic interaction is required. This work proposes an automated method for extraction of knowledge from National Comprehensive Cancer Network (NCCN) CPGs in Oncology and generating a structured model containing the retrieved knowledge. The proposed method was tested using two versions of NCCN Non-Small Cell Lung Cancer (NSCLC) CPG to demonstrate the effectiveness in faithful extraction and modeling of knowledge. Three enrichment strategies using Cancer staging information, Unified Medical Language System (UMLS) Metathesaurus & National Cancer Institute thesaurus (NCIt) concepts, and Node classification are also presented to enhance the model towards enabling programmatic traversal and querying of cancer care guidelines. The Node classification was performed using a Support Vector Machine (SVM) model, achieving a classification accuracy of 0.81 with 10-fold cross-validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge