Arunima Sarkar
AD-DAE: Unsupervised Modeling of Longitudinal Alzheimer's Disease Progression with Diffusion Auto-Encoder
Nov 08, 2025Abstract:Generative modeling frameworks have emerged as an effective approach to capture high-dimensional image distributions from large datasets without requiring domain-specific knowledge, a capability essential for longitudinal disease progression modeling. Recent generative modeling approaches have attempted to capture progression by mapping images into a latent representational space and then controlling and guiding the representations to generate follow-up images from a baseline image. However, existing approaches impose constraints on distribution learning, leading to latent spaces with limited controllability to generate follow-up images without explicit supervision from subject-specific longitudinal images. In order to enable controlled movements in the latent representational space and generate progression images from a baseline image in an unsupervised manner, we introduce a conditionable Diffusion Auto-encoder framework. The explicit encoding mechanism of image-diffusion auto-encoders forms a compact latent space capturing high-level semantics, providing means to disentangle information relevant for progression. Our approach leverages this latent space to condition and apply controlled shifts to baseline representations for generating follow-up. Controllability is induced by restricting these shifts to a subspace, thereby isolating progression-related factors from subject identity-preserving components. The shifts are implicitly guided by correlating with progression attributes, without requiring subject-specific longitudinal supervision. We validate the generations through image quality metrics, volumetric progression analysis, and downstream classification in Alzheimer's disease datasets from two different sources and disease categories. This demonstrates the effectiveness of our approach for Alzheimer's progression modeling and longitudinal image generation.
DCE-FORMER: A Transformer-based Model With Mutual Information And Frequency-based Loss Functions For Early And Late Response Prediction In Prostate DCE-MRI
Feb 03, 2024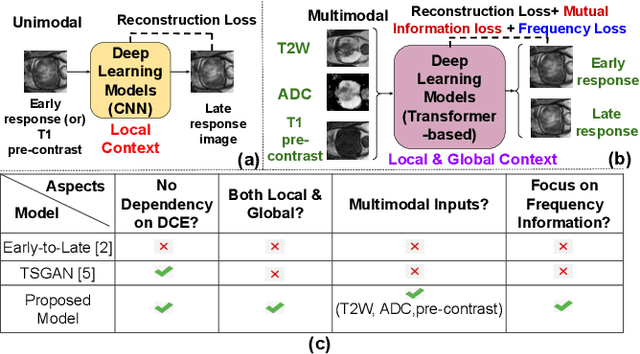
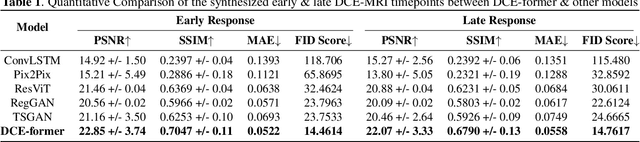
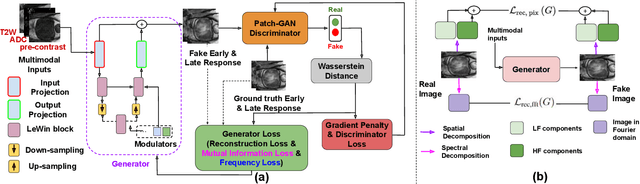
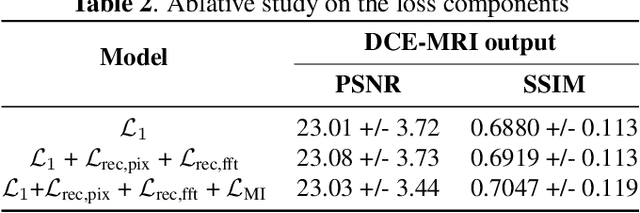
Abstract:Dynamic Contrast Enhanced Magnetic Resonance Imaging aids in the detection and assessment of tumor aggressiveness by using a Gadolinium-based contrast agent (GBCA). However, GBCA is known to have potential toxic effects. This risk can be avoided if we obtain DCE-MRI images without using GBCA. We propose, DCE-former, a transformer-based neural network to generate early and late response prostate DCE-MRI images from non-contrast multimodal inputs (T2 weighted, Apparent Diffusion Coefficient, and T1 pre-contrast MRI). Additionally, we introduce (i) a mutual information loss function to capture the complementary information about contrast uptake, and (ii) a frequency-based loss function in the pixel and Fourier space to learn local and global hyper-intensity patterns in DCE-MRI. Extensive experiments show that DCE-former outperforms other methods with improvement margins of +1.39 dB and +1.19 db in PSNR, +0.068 and +0.055 in SSIM, and -0.012 and -0.013 in Mean Absolute Error for early and late response DCE-MRI, respectively.
Automated Knowledge Modeling for Cancer Clinical Practice Guidelines
Jul 15, 2023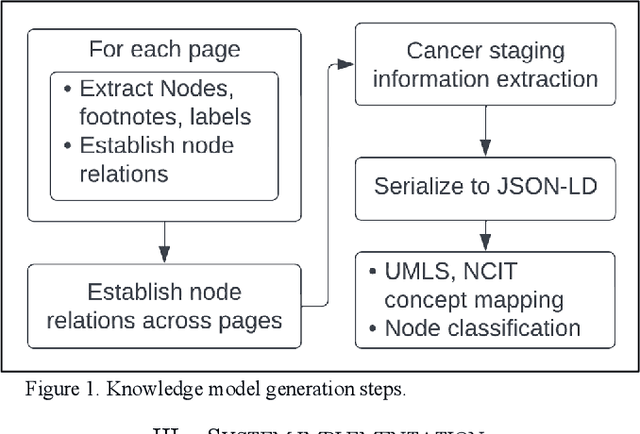
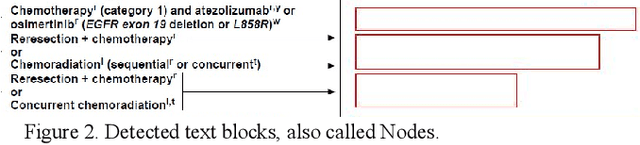
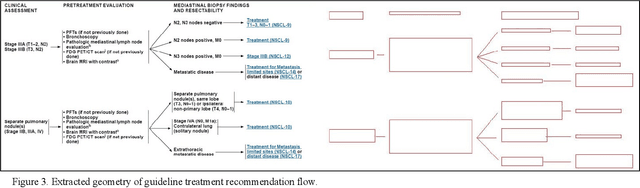
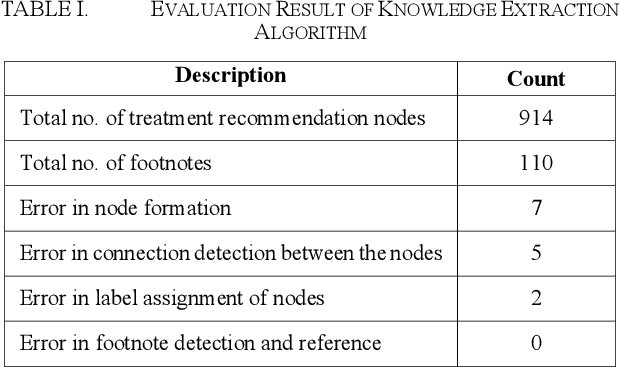
Abstract:Clinical Practice Guidelines (CPGs) for cancer diseases evolve rapidly due to new evidence generated by active research. Currently, CPGs are primarily published in a document format that is ill-suited for managing this developing knowledge. A knowledge model of the guidelines document suitable for programmatic interaction is required. This work proposes an automated method for extraction of knowledge from National Comprehensive Cancer Network (NCCN) CPGs in Oncology and generating a structured model containing the retrieved knowledge. The proposed method was tested using two versions of NCCN Non-Small Cell Lung Cancer (NSCLC) CPG to demonstrate the effectiveness in faithful extraction and modeling of knowledge. Three enrichment strategies using Cancer staging information, Unified Medical Language System (UMLS) Metathesaurus & National Cancer Institute thesaurus (NCIt) concepts, and Node classification are also presented to enhance the model towards enabling programmatic traversal and querying of cancer care guidelines. The Node classification was performed using a Support Vector Machine (SVM) model, achieving a classification accuracy of 0.81 with 10-fold cross-validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge