Teng Xiao
Olmo 3
Dec 15, 2025Abstract:We introduce Olmo 3, a family of state-of-the-art, fully-open language models at the 7B and 32B parameter scales. Olmo 3 model construction targets long-context reasoning, function calling, coding, instruction following, general chat, and knowledge recall. This release includes the entire model flow, i.e., the full lifecycle of the family of models, including every stage, checkpoint, data point, and dependency used to build it. Our flagship model, Olmo 3 Think 32B, is the strongest fully-open thinking model released to-date.
Simple Denoising Diffusion Language Models
Oct 27, 2025Abstract:Diffusion models have recently been extended to language generation through Masked Diffusion Language Models (MDLMs), which achieve performance competitive with strong autoregressive models. However, MDLMs tend to degrade in the few-step regime and cannot directly adopt existing few-step distillation methods designed for continuous diffusion models, as they lack the intrinsic property of mapping from noise to data. Recent Uniform-state Diffusion Models (USDMs), initialized from a uniform prior, alleviate some limitations but still suffer from complex loss formulations that hinder scalability. In this work, we propose a simplified denoising-based loss for USDMs that optimizes only noise-replaced tokens, stabilizing training and matching ELBO-level performance. Furthermore, by framing denoising as self-supervised learning, we introduce a simple modification to our denoising loss with contrastive-inspired negative gradients, which is practical and yield additional improvements in generation quality.
Incentivizing Strong Reasoning from Weak Supervision
May 28, 2025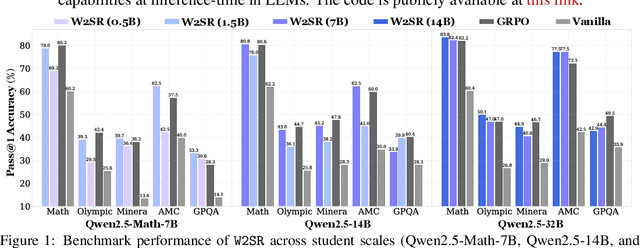
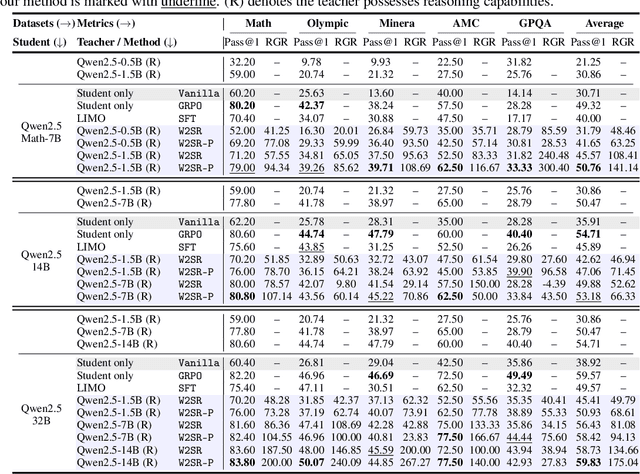
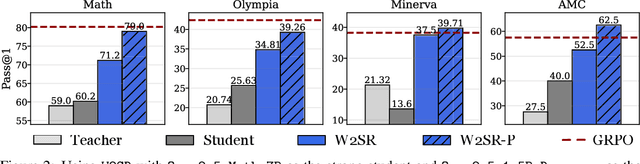
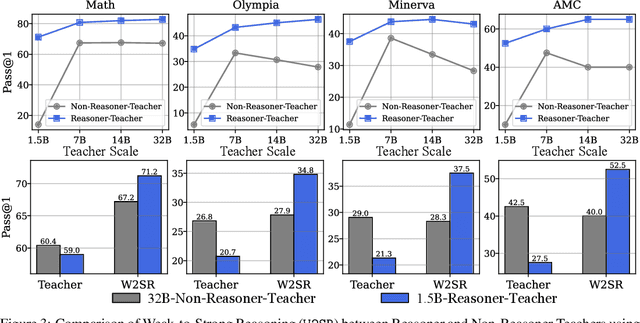
Abstract:Large language models (LLMs) have demonstrated impressive performance on reasoning-intensive tasks, but enhancing their reasoning abilities typically relies on either reinforcement learning (RL) with verifiable signals or supervised fine-tuning (SFT) with high-quality long chain-of-thought (CoT) demonstrations, both of which are expensive. In this paper, we study a novel problem of incentivizing the reasoning capacity of LLMs without expensive high-quality demonstrations and reinforcement learning. We investigate whether the reasoning capabilities of LLMs can be effectively incentivized via supervision from significantly weaker models. We further analyze when and why such weak supervision succeeds in eliciting reasoning abilities in stronger models. Our findings show that supervision from significantly weaker reasoners can substantially improve student reasoning performance, recovering close to 94% of the gains of expensive RL at a fraction of the cost. Experiments across diverse benchmarks and model architectures demonstrate that weak reasoners can effectively incentivize reasoning in stronger student models, consistently improving performance across a wide range of reasoning tasks. Our results suggest that this simple weak-to-strong paradigm is a promising and generalizable alternative to costly methods for incentivizing strong reasoning capabilities at inference-time in LLMs. The code is publicly available at https://github.com/yuanyige/w2sr.
Inference-time Alignment in Continuous Space
May 26, 2025Abstract:Aligning large language models with human feedback at inference time has received increasing attention due to its flexibility. Existing methods rely on generating multiple responses from the base policy for search using a reward model, which can be considered as searching in a discrete response space. However, these methods struggle to explore informative candidates when the base policy is weak or the candidate set is small, resulting in limited effectiveness. In this paper, to address this problem, we propose Simple Energy Adaptation ($\textbf{SEA}$), a simple yet effective algorithm for inference-time alignment. In contrast to expensive search over the discrete space, SEA directly adapts original responses from the base policy toward the optimal one via gradient-based sampling in continuous latent space. Specifically, SEA formulates inference as an iterative optimization procedure on an energy function over actions in the continuous space defined by the optimal policy, enabling simple and effective alignment. For instance, despite its simplicity, SEA outperforms the second-best baseline with a relative improvement of up to $ \textbf{77.51%}$ on AdvBench and $\textbf{16.36%}$ on MATH. Our code is publicly available at https://github.com/yuanyige/SEA
Incentivizing Reasoning from Weak Supervision
May 26, 2025



Abstract:Large language models (LLMs) have demonstrated impressive performance on reasoning-intensive tasks, but enhancing their reasoning abilities typically relies on either reinforcement learning (RL) with verifiable signals or supervised fine-tuning (SFT) with high-quality long chain-of-thought (CoT) demonstrations, both of which are expensive. In this paper, we study a novel problem of incentivizing the reasoning capacity of LLMs without expensive high-quality demonstrations and reinforcement learning. We investigate whether the reasoning capabilities of LLMs can be effectively incentivized via supervision from significantly weaker models. We further analyze when and why such weak supervision succeeds in eliciting reasoning abilities in stronger models. Our findings show that supervision from significantly weaker reasoners can substantially improve student reasoning performance, recovering close to 94% of the gains of expensive RL at a fraction of the cost. Experiments across diverse benchmarks and model architectures demonstrate that weak reasoners can effectively incentivize reasoning in stronger student models, consistently improving performance across a wide range of reasoning tasks. Our results suggest that this simple weak-to-strong paradigm is a promising and generalizable alternative to costly methods for incentivizing strong reasoning capabilities at inference-time in LLMs. The code is publicly available at https://github.com/yuanyige/W2SR.
InfoPO: On Mutual Information Maximization for Large Language Model Alignment
May 13, 2025Abstract:We study the post-training of large language models (LLMs) with human preference data. Recently, direct preference optimization and its variants have shown considerable promise in aligning language models, eliminating the need for reward models and online sampling. Despite these benefits, these methods rely on explicit assumptions about the Bradley-Terry (BT) model, which makes them prone to overfitting and results in suboptimal performance, particularly on reasoning-heavy tasks. To address these challenges, we propose a principled preference fine-tuning algorithm called InfoPO, which effectively and efficiently aligns large language models using preference data. InfoPO eliminates the reliance on the BT model and prevents the likelihood of the chosen response from decreasing. Extensive experiments confirm that InfoPO consistently outperforms established baselines on widely used open benchmarks, particularly in reasoning tasks.
A Deep Single Image Rectification Approach for Pan-Tilt-Zoom Cameras
Apr 09, 2025Abstract:Pan-Tilt-Zoom (PTZ) cameras with wide-angle lenses are widely used in surveillance but often require image rectification due to their inherent nonlinear distortions. Current deep learning approaches typically struggle to maintain fine-grained geometric details, resulting in inaccurate rectification. This paper presents a Forward Distortion and Backward Warping Network (FDBW-Net), a novel framework for wide-angle image rectification. It begins by using a forward distortion model to synthesize barrel-distorted images, reducing pixel redundancy and preventing blur. The network employs a pyramid context encoder with attention mechanisms to generate backward warping flows containing geometric details. Then, a multi-scale decoder is used to restore distorted features and output rectified images. FDBW-Net's performance is validated on diverse datasets: public benchmarks, AirSim-rendered PTZ camera imagery, and real-scene PTZ camera datasets. It demonstrates that FDBW-Net achieves SOTA performance in distortion rectification, boosting the adaptability of PTZ cameras for practical visual applications.
On a Connection Between Imitation Learning and RLHF
Mar 07, 2025Abstract:This work studies the alignment of large language models with preference data from an imitation learning perspective. We establish a close theoretical connection between reinforcement learning from human feedback RLHF and imitation learning (IL), revealing that RLHF implicitly performs imitation learning on the preference data distribution. Building on this connection, we propose DIL, a principled framework that directly optimizes the imitation learning objective. DIL provides a unified imitation learning perspective on alignment, encompassing existing alignment algorithms as special cases while naturally introducing new variants. By bridging IL and RLHF, DIL offers new insights into alignment with RLHF. Extensive experiments demonstrate that DIL outperforms existing methods on various challenging benchmarks.
SimPER: A Minimalist Approach to Preference Alignment without Hyperparameters
Feb 04, 2025



Abstract:Existing preference optimization objectives for language model alignment require additional hyperparameters that must be extensively tuned to achieve optimal performance, increasing both the complexity and time required for fine-tuning large language models. In this paper, we propose a simple yet effective hyperparameter-free preference optimization algorithm for alignment. We observe that promising performance can be achieved simply by optimizing inverse perplexity, which is calculated as the inverse of the exponentiated average log-likelihood of the chosen and rejected responses in the preference dataset. The resulting simple learning objective, SimPER, is easy to implement and eliminates the need for expensive hyperparameter tuning and a reference model, making it both computationally and memory efficient. Extensive experiments on widely used real-world benchmarks, including MT-Bench, AlpacaEval 2, and 10 key benchmarks of the Open LLM Leaderboard with 5 base models, demonstrate that SimPER consistently and significantly outperforms existing approaches-even without any hyperparameters or a reference model . For example, despite its simplicity, SimPER outperforms state-of-the-art methods by up to 5.7 points on AlpacaEval 2 and achieves the highest average ranking across 10 benchmarks on the Open LLM Leaderboard. The source code for SimPER is publicly available at: https://github.com/tengxiao1/SimPER.
Cal-DPO: Calibrated Direct Preference Optimization for Language Model Alignment
Dec 19, 2024Abstract:We study the problem of aligning large language models (LLMs) with human preference data. Contrastive preference optimization has shown promising results in aligning LLMs with available preference data by optimizing the implicit reward associated with the policy. However, the contrastive objective focuses mainly on the relative values of implicit rewards associated with two responses while ignoring their actual values, resulting in suboptimal alignment with human preferences. To address this limitation, we propose calibrated direct preference optimization (Cal-DPO), a simple yet effective algorithm. We show that substantial improvement in alignment with the given preferences can be achieved simply by calibrating the implicit reward to ensure that the learned implicit rewards are comparable in scale to the ground-truth rewards. We demonstrate the theoretical advantages of Cal-DPO over existing approaches. The results of our experiments on a variety of standard benchmarks show that Cal-DPO remarkably improves off-the-shelf methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge