Vasant G Honavar
On a Connection Between Imitation Learning and RLHF
Mar 07, 2025Abstract:This work studies the alignment of large language models with preference data from an imitation learning perspective. We establish a close theoretical connection between reinforcement learning from human feedback RLHF and imitation learning (IL), revealing that RLHF implicitly performs imitation learning on the preference data distribution. Building on this connection, we propose DIL, a principled framework that directly optimizes the imitation learning objective. DIL provides a unified imitation learning perspective on alignment, encompassing existing alignment algorithms as special cases while naturally introducing new variants. By bridging IL and RLHF, DIL offers new insights into alignment with RLHF. Extensive experiments demonstrate that DIL outperforms existing methods on various challenging benchmarks.
SimPER: A Minimalist Approach to Preference Alignment without Hyperparameters
Feb 04, 2025



Abstract:Existing preference optimization objectives for language model alignment require additional hyperparameters that must be extensively tuned to achieve optimal performance, increasing both the complexity and time required for fine-tuning large language models. In this paper, we propose a simple yet effective hyperparameter-free preference optimization algorithm for alignment. We observe that promising performance can be achieved simply by optimizing inverse perplexity, which is calculated as the inverse of the exponentiated average log-likelihood of the chosen and rejected responses in the preference dataset. The resulting simple learning objective, SimPER, is easy to implement and eliminates the need for expensive hyperparameter tuning and a reference model, making it both computationally and memory efficient. Extensive experiments on widely used real-world benchmarks, including MT-Bench, AlpacaEval 2, and 10 key benchmarks of the Open LLM Leaderboard with 5 base models, demonstrate that SimPER consistently and significantly outperforms existing approaches-even without any hyperparameters or a reference model . For example, despite its simplicity, SimPER outperforms state-of-the-art methods by up to 5.7 points on AlpacaEval 2 and achieves the highest average ranking across 10 benchmarks on the Open LLM Leaderboard. The source code for SimPER is publicly available at: https://github.com/tengxiao1/SimPER.
Cal-DPO: Calibrated Direct Preference Optimization for Language Model Alignment
Dec 19, 2024Abstract:We study the problem of aligning large language models (LLMs) with human preference data. Contrastive preference optimization has shown promising results in aligning LLMs with available preference data by optimizing the implicit reward associated with the policy. However, the contrastive objective focuses mainly on the relative values of implicit rewards associated with two responses while ignoring their actual values, resulting in suboptimal alignment with human preferences. To address this limitation, we propose calibrated direct preference optimization (Cal-DPO), a simple yet effective algorithm. We show that substantial improvement in alignment with the given preferences can be achieved simply by calibrating the implicit reward to ensure that the learned implicit rewards are comparable in scale to the ground-truth rewards. We demonstrate the theoretical advantages of Cal-DPO over existing approaches. The results of our experiments on a variety of standard benchmarks show that Cal-DPO remarkably improves off-the-shelf methods.
How to Leverage Demonstration Data in Alignment for Large Language Model? A Self-Imitation Learning Perspective
Oct 14, 2024Abstract:This paper introduces a novel generalized self-imitation learning ($\textbf{GSIL}$) framework, which effectively and efficiently aligns large language models with offline demonstration data. We develop $\textbf{GSIL}$ by deriving a surrogate objective of imitation learning with density ratio estimates, facilitating the use of self-generated data and optimizing the imitation learning objective with simple classification losses. $\textbf{GSIL}$ eliminates the need for complex adversarial training in standard imitation learning, achieving lightweight and efficient fine-tuning for large language models. In addition, $\textbf{GSIL}$ encompasses a family of offline losses parameterized by a general class of convex functions for density ratio estimation and enables a unified view for alignment with demonstration data. Extensive experiments show that $\textbf{GSIL}$ consistently and significantly outperforms baselines in many challenging benchmarks, such as coding (HuamnEval), mathematical reasoning (GSM8K) and instruction-following benchmark (MT-Bench).
3M-Diffusion: Latent Multi-Modal Diffusion for Text-Guided Generation of Molecular Graphs
Mar 11, 2024



Abstract:Generating molecules with desired properties is a critical task with broad applications in drug discovery and materials design. Inspired by recent advances in large language models, there is a growing interest in using natural language descriptions of molecules to generate molecules with the desired properties. Most existing methods focus on generating molecules that precisely match the text description. However, practical applications call for methods that generate diverse, and ideally novel, molecules with the desired properties. We propose 3M-Diffusion, a novel multi-modal molecular graph generation method, to address this challenge. 3M-Diffusion first encodes molecular graphs into a graph latent space aligned with text descriptions. It then reconstructs the molecular structure and atomic attributes based on the given text descriptions using the molecule decoder. It then learns a probabilistic mapping from the text space to the latent molecular graph space using a diffusion model. The results of our extensive experiments on several datasets demonstrate that 3M-Diffusion can generate high-quality, novel and diverse molecular graphs that semantically match the textual description provided.
EsaCL: Efficient Continual Learning of Sparse Models
Jan 11, 2024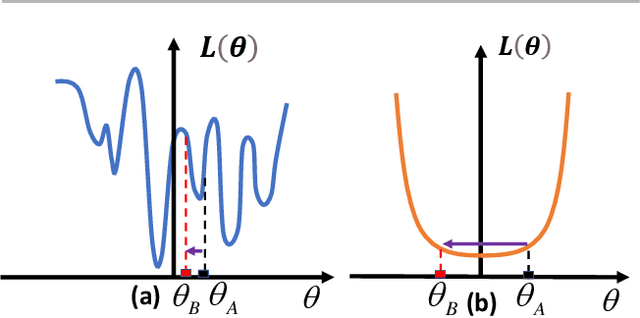
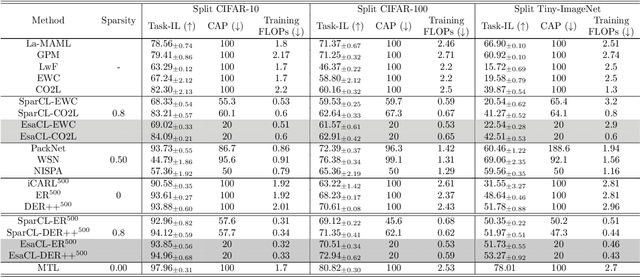

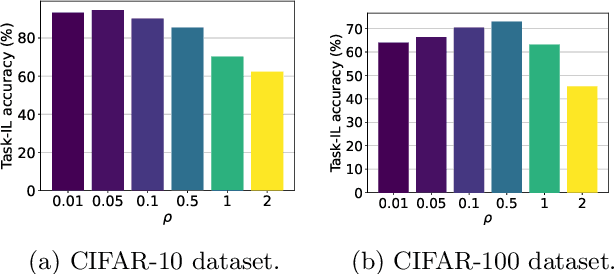
Abstract:A key challenge in the continual learning setting is to efficiently learn a sequence of tasks without forgetting how to perform previously learned tasks. Many existing approaches to this problem work by either retraining the model on previous tasks or by expanding the model to accommodate new tasks. However, these approaches typically suffer from increased storage and computational requirements, a problem that is worsened in the case of sparse models due to need for expensive re-training after sparsification. To address this challenge, we propose a new method for efficient continual learning of sparse models (EsaCL) that can automatically prune redundant parameters without adversely impacting the model's predictive power, and circumvent the need of retraining. We conduct a theoretical analysis of loss landscapes with parameter pruning, and design a directional pruning (SDP) strategy that is informed by the sharpness of the loss function with respect to the model parameters. SDP ensures model with minimal loss of predictive accuracy, accelerating the learning of sparse models at each stage. To accelerate model update, we introduce an intelligent data selection (IDS) strategy that can identify critical instances for estimating loss landscape, yielding substantially improved data efficiency. The results of our experiments show that EsaCL achieves performance that is competitive with the state-of-the-art methods on three continual learning benchmarks, while using substantially reduced memory and computational resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge