Rizwan Qureshi
Foundation Models in Biomedical Imaging: Turning Hype into Reality
Dec 17, 2025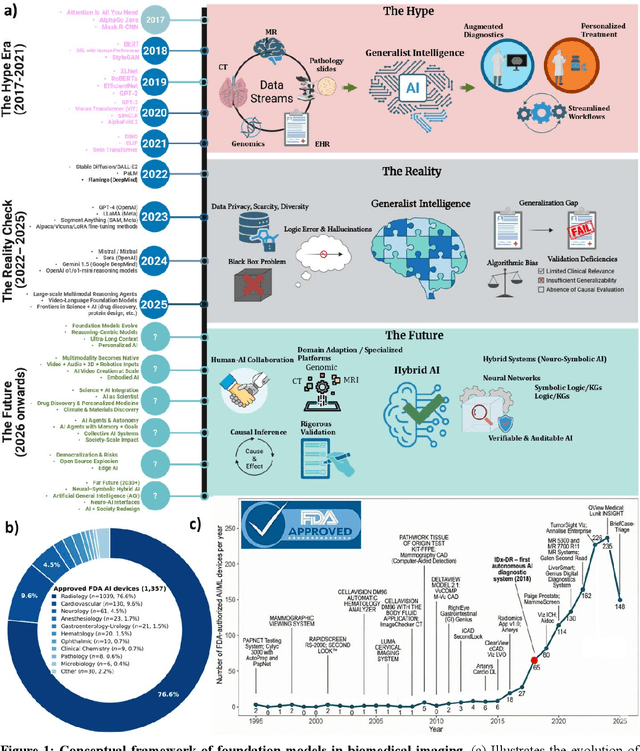
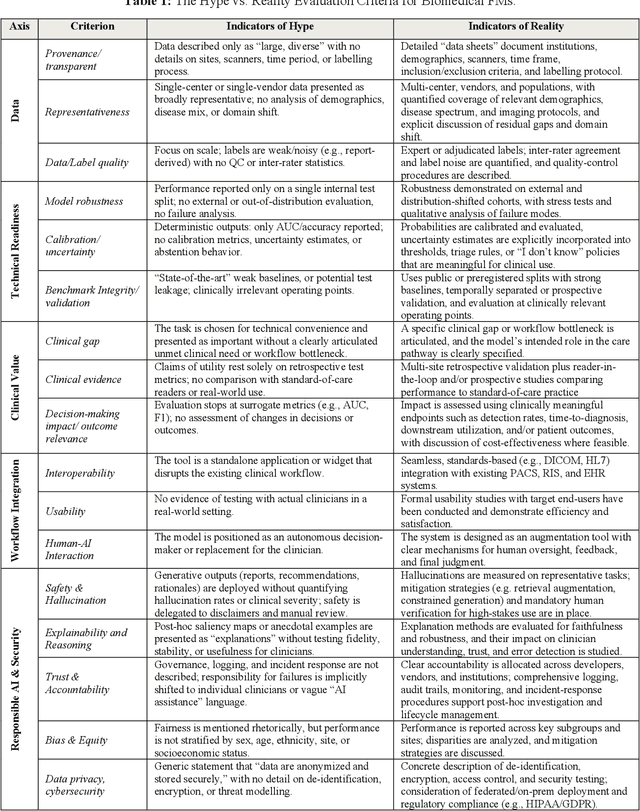
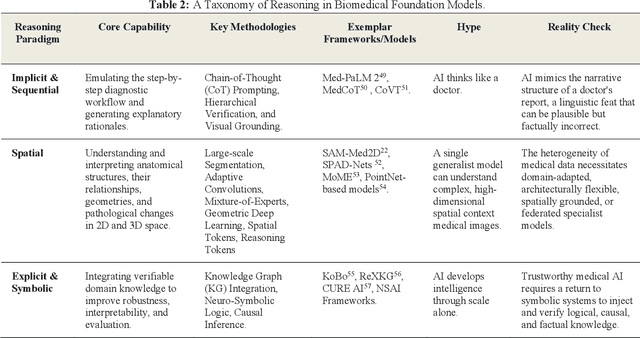
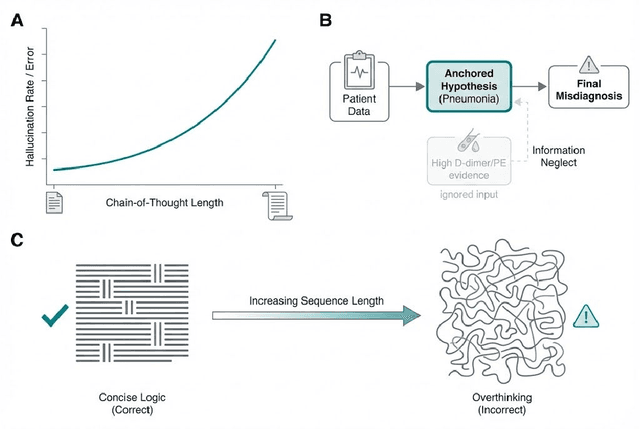
Abstract:Foundation models (FMs) are driving a prominent shift in artificial intelligence across different domains, including biomedical imaging. These models are designed to move beyond narrow pattern recognition towards emulating sophisticated clinical reasoning, understanding complex spatial relationships, and integrating multimodal data with unprecedented flexibility. However, a critical gap exists between this potential and the current reality, where the clinical evaluation and deployment of FMs are hampered by significant challenges. Herein, we critically assess the current state-of-the-art, analyzing hype by examining the core capabilities and limitations of FMs in the biomedical domain. We also provide a taxonomy of reasoning, ranging from emulated sequential logic and spatial understanding to the integration of explicit symbolic knowledge, to evaluate whether these models exhibit genuine cognition or merely mimic surface-level patterns. We argue that a critical frontier lies beyond statistical correlation, in the pursuit of causal inference, which is essential for building robust models that understand cause and effect. Furthermore, we discuss the paramount issues in deployment stemming from trustworthiness, bias, and safety, dissecting the challenges of algorithmic bias, data bias and privacy, and model hallucinations. We also draw attention to the need for more inclusive, rigorous, and clinically relevant validation frameworks to ensure their safe and ethical application. We conclude that while the vision of autonomous AI-doctors remains distant, the immediate reality is the emergence of powerful technology and assistive tools that would benefit clinical practice. The future of FMs in biomedical imaging hinges not on scale alone, but on developing hybrid, causally aware, and verifiably safe systems that augment, rather than replace, human expertise.
Thinking Beyond Tokens: From Brain-Inspired Intelligence to Cognitive Foundations for Artificial General Intelligence and its Societal Impact
Jul 01, 2025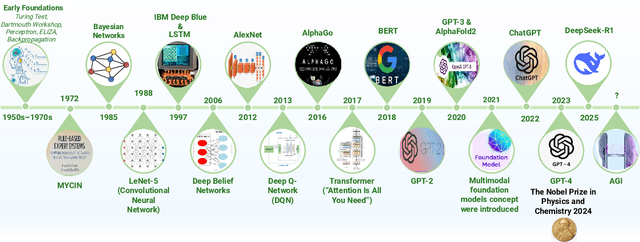
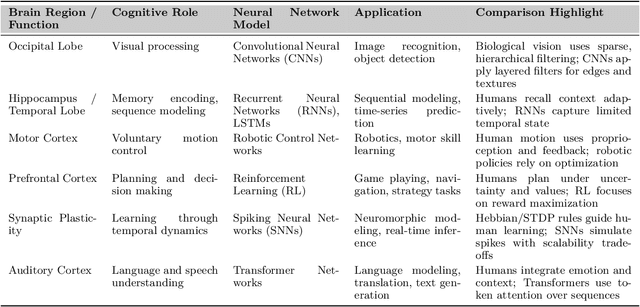
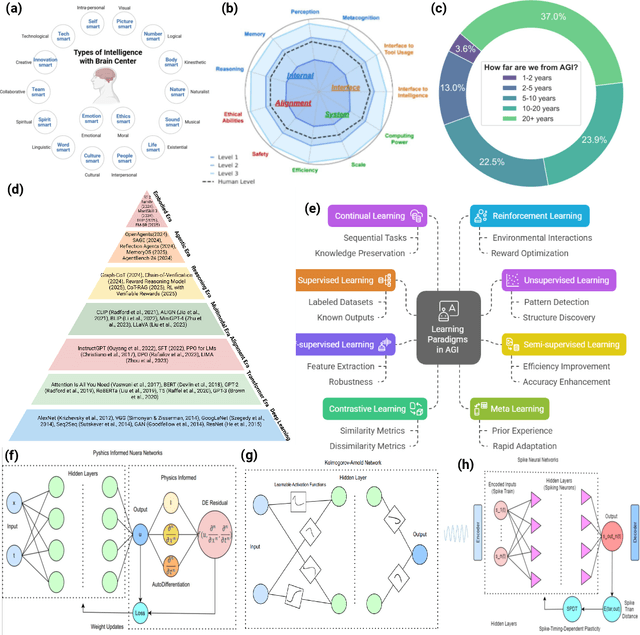
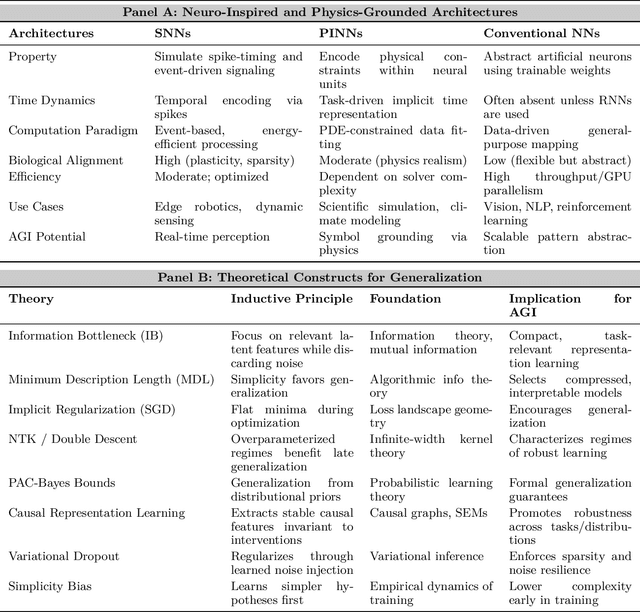
Abstract:Can machines truly think, reason and act in domains like humans? This enduring question continues to shape the pursuit of Artificial General Intelligence (AGI). Despite the growing capabilities of models such as GPT-4.5, DeepSeek, Claude 3.5 Sonnet, Phi-4, and Grok 3, which exhibit multimodal fluency and partial reasoning, these systems remain fundamentally limited by their reliance on token-level prediction and lack of grounded agency. This paper offers a cross-disciplinary synthesis of AGI development, spanning artificial intelligence, cognitive neuroscience, psychology, generative models, and agent-based systems. We analyze the architectural and cognitive foundations of general intelligence, highlighting the role of modular reasoning, persistent memory, and multi-agent coordination. In particular, we emphasize the rise of Agentic RAG frameworks that combine retrieval, planning, and dynamic tool use to enable more adaptive behavior. We discuss generalization strategies, including information compression, test-time adaptation, and training-free methods, as critical pathways toward flexible, domain-agnostic intelligence. Vision-Language Models (VLMs) are reexamined not just as perception modules but as evolving interfaces for embodied understanding and collaborative task completion. We also argue that true intelligence arises not from scale alone but from the integration of memory and reasoning: an orchestration of modular, interactive, and self-improving components where compression enables adaptive behavior. Drawing on advances in neurosymbolic systems, reinforcement learning, and cognitive scaffolding, we explore how recent architectures begin to bridge the gap between statistical learning and goal-directed cognition. Finally, we identify key scientific, technical, and ethical challenges on the path to AGI.
Leveraging MIMIC Datasets for Better Digital Health: A Review on Open Problems, Progress Highlights, and Future Promises
Jun 15, 2025Abstract:The Medical Information Mart for Intensive Care (MIMIC) datasets have become the Kernel of Digital Health Research by providing freely accessible, deidentified records from tens of thousands of critical care admissions, enabling a broad spectrum of applications in clinical decision support, outcome prediction, and healthcare analytics. Although numerous studies and surveys have explored the predictive power and clinical utility of MIMIC based models, critical challenges in data integration, representation, and interoperability remain underexplored. This paper presents a comprehensive survey that focuses uniquely on open problems. We identify persistent issues such as data granularity, cardinality limitations, heterogeneous coding schemes, and ethical constraints that hinder the generalizability and real-time implementation of machine learning models. We highlight key progress in dimensionality reduction, temporal modelling, causal inference, and privacy preserving analytics, while also outlining promising directions including hybrid modelling, federated learning, and standardized preprocessing pipelines. By critically examining these structural limitations and their implications, this survey offers actionable insights to guide the next generation of MIMIC powered digital health innovations.
A Layered Self-Supervised Knowledge Distillation Framework for Efficient Multimodal Learning on the Edge
Jun 08, 2025Abstract:We introduce Layered Self-Supervised Knowledge Distillation (LSSKD) framework for training compact deep learning models. Unlike traditional methods that rely on pre-trained teacher networks, our approach appends auxiliary classifiers to intermediate feature maps, generating diverse self-supervised knowledge and enabling one-to-one transfer across different network stages. Our method achieves an average improvement of 4.54\% over the state-of-the-art PS-KD method and a 1.14% gain over SSKD on CIFAR-100, with a 0.32% improvement on ImageNet compared to HASSKD. Experiments on Tiny ImageNet and CIFAR-100 under few-shot learning scenarios also achieve state-of-the-art results. These findings demonstrate the effectiveness of our approach in enhancing model generalization and performance without the need for large over-parameterized teacher networks. Importantly, at the inference stage, all auxiliary classifiers can be removed, yielding no extra computational cost. This makes our model suitable for deploying small language models on affordable low-computing devices. Owing to its lightweight design and adaptability, our framework is particularly suitable for multimodal sensing and cyber-physical environments that require efficient and responsive inference. LSSKD facilitates the development of intelligent agents capable of learning from limited sensory data under weak supervision.
SatelliteFormula: Multi-Modal Symbolic Regression from Remote Sensing Imagery for Physics Discovery
Jun 06, 2025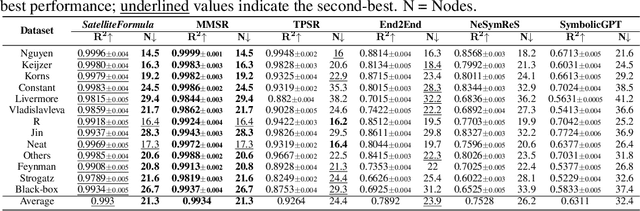
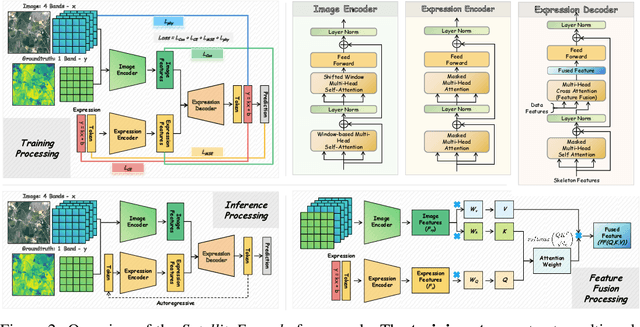
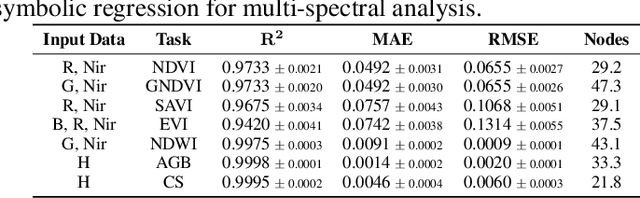
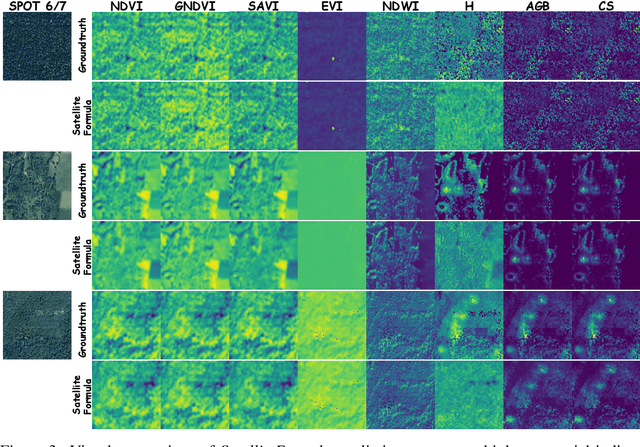
Abstract:We propose SatelliteFormula, a novel symbolic regression framework that derives physically interpretable expressions directly from multi-spectral remote sensing imagery. Unlike traditional empirical indices or black-box learning models, SatelliteFormula combines a Vision Transformer-based encoder for spatial-spectral feature extraction with physics-guided constraints to ensure consistency and interpretability. Existing symbolic regression methods struggle with the high-dimensional complexity of multi-spectral data; our method addresses this by integrating transformer representations into a symbolic optimizer that balances accuracy and physical plausibility. Extensive experiments on benchmark datasets and remote sensing tasks demonstrate superior performance, stability, and generalization compared to state-of-the-art baselines. SatelliteFormula enables interpretable modeling of complex environmental variables, bridging the gap between data-driven learning and physical understanding.
Just as Humans Need Vaccines, So Do Models: Model Immunization to Combat Falsehoods
May 23, 2025Abstract:Generative AI models often learn and reproduce false information present in their training corpora. This position paper argues that, analogous to biological immunization, where controlled exposure to a weakened pathogen builds immunity, AI models should be fine tuned on small, quarantined sets of explicitly labeled falsehoods as a "vaccine" against misinformation. These curated false examples are periodically injected during finetuning, strengthening the model ability to recognize and reject misleading claims while preserving accuracy on truthful inputs. An illustrative case study shows that immunized models generate substantially less misinformation than baselines. To our knowledge, this is the first training framework that treats fact checked falsehoods themselves as a supervised vaccine, rather than relying on input perturbations or generic human feedback signals, to harden models against future misinformation. We also outline ethical safeguards and governance controls to ensure the safe use of false data. Model immunization offers a proactive paradigm for aligning AI systems with factuality.
VLDBench: Vision Language Models Disinformation Detection Benchmark
Feb 17, 2025



Abstract:The rapid rise of AI-generated content has made detecting disinformation increasingly challenging. In particular, multimodal disinformation, i.e., online posts-articles that contain images and texts with fabricated information are specially designed to deceive. While existing AI safety benchmarks primarily address bias and toxicity, multimodal disinformation detection remains largely underexplored. To address this challenge, we present the Vision-Language Disinformation Detection Benchmark VLDBench, the first comprehensive benchmark for detecting disinformation across both unimodal (text-only) and multimodal (text and image) content, comprising 31,000} news article-image pairs, spanning 13 distinct categories, for robust evaluation. VLDBench features a rigorous semi-automated data curation pipeline, with 22 domain experts dedicating 300 plus hours} to annotation, achieving a strong inter-annotator agreement (Cohen kappa = 0.78). We extensively evaluate state-of-the-art Large Language Models (LLMs) and Vision-Language Models (VLMs), demonstrating that integrating textual and visual cues in multimodal news posts improves disinformation detection accuracy by 5 - 35 % compared to unimodal models. Developed in alignment with AI governance frameworks such as the EU AI Act, NIST guidelines, and the MIT AI Risk Repository 2024, VLDBench is expected to become a benchmark for detecting disinformation in online multi-modal contents. Our code and data will be publicly available.
Technical note on calibrating vision-language models under covariate shift
Feb 11, 2025



Abstract:Despite being a successful example of emerging capability, vision-language foundation models for low-shot vision classification have a limited ability to sufficiently generalize to the target data distribution due to sample poverty, leading to sensitivity to variations in the data. A popular mitigation strategy is finetuning over multiple datasets, but domain generalization is expensive when practiced in this manner. This work examines both covariate shift between pre-training data and the underspecified target data, and \textit{confidence misalignment}, where the model's prediction confidence amplified by the limited data availability. We propose \textit{Confidence-Calibrated Covariate Shift Correction ($C3SC$)}, a unified framework to mitigate both covariate shift and confidence misalignment. $C3SC$ leverages Fisher information penalty for covariate shift correction and confidence misalignment penalty (CMP) to lower confidence on misclassified examples. Experimental results across various vision and covariate shift datasets demonstrates that $C3SC$ significantly improves in calibration (ECE) by $5.82\%$ at maximum. $C3SC$ shows better robustness as well by showing $3.5\%$ improvement in accuracy metric on challenging covariate shift datasets, making $C3SC$ a promising solution for reliable real-world vision-language low-shot applications under distribution shift.
Voxel-Aggergated Feature Synthesis: Efficient Dense Mapping for Simulated 3D Reasoning
Nov 15, 2024



Abstract:We address the issue of the exploding computational requirements of recent State-of-the-art (SOTA) open set multimodel 3D mapping (dense 3D mapping) algorithms and present Voxel-Aggregated Feature Synthesis (VAFS), a novel approach to dense 3D mapping in simulation. Dense 3D mapping involves segmenting and embedding sequential RGBD frames which are then fused into 3D. This leads to redundant computation as the differences between frames are small but all are individually segmented and embedded. This makes dense 3D mapping impractical for research involving embodied agents in which the environment, and thus the mapping, must be modified with regularity. VAFS drastically reduces this computation by using the segmented point cloud computed by a simulator's physics engine and synthesizing views of each region. This reduces the number of features to embed from the number of captured RGBD frames to the number of objects in the scene, effectively allowing a "ground truth" semantic map to be computed an order of magnitude faster than traditional methods. We test the resulting representation by assessing the IoU scores of semantic queries for different objects in the simulated scene, and find that VAFS exceeds the accuracy and speed of prior dense 3D mapping techniques.
Brain Tumor Synthetic Data Generation with Adaptive StyleGANs
Dec 04, 2022Abstract:Generative models have been very successful over the years and have received significant attention for synthetic data generation. As deep learning models are getting more and more complex, they require large amounts of data to perform accurately. In medical image analysis, such generative models play a crucial role as the available data is limited due to challenges related to data privacy, lack of data diversity, or uneven data distributions. In this paper, we present a method to generate brain tumor MRI images using generative adversarial networks. We have utilized StyleGAN2 with ADA methodology to generate high-quality brain MRI with tumors while using a significantly smaller amount of training data when compared to the existing approaches. We use three pre-trained models for transfer learning. Results demonstrate that the proposed method can learn the distributions of brain tumors. Furthermore, the model can generate high-quality synthetic brain MRI with a tumor that can limit the small sample size issues. The approach can addresses the limited data availability by generating realistic-looking brain MRI with tumors. The code is available at: ~\url{https://github.com/rizwanqureshi123/Brain-Tumor-Synthetic-Data}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge