Hazrat Ali
Foundation Models in Biomedical Imaging: Turning Hype into Reality
Dec 17, 2025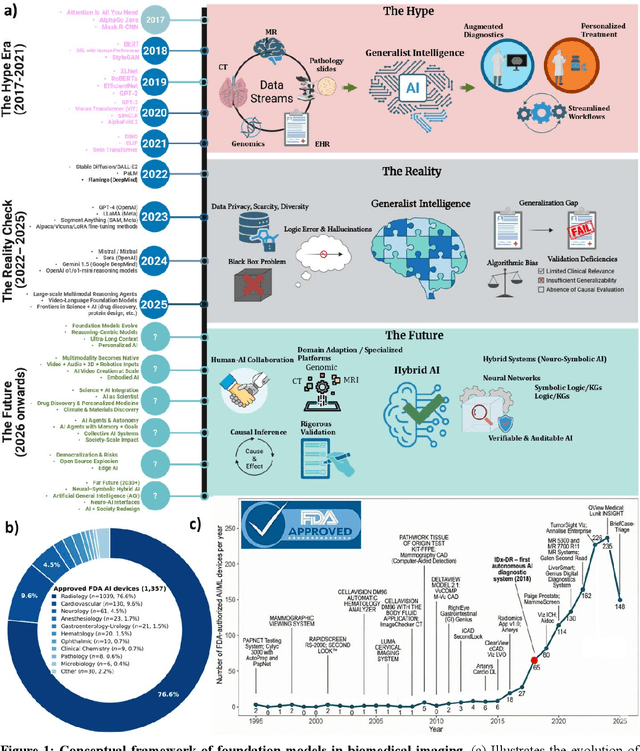
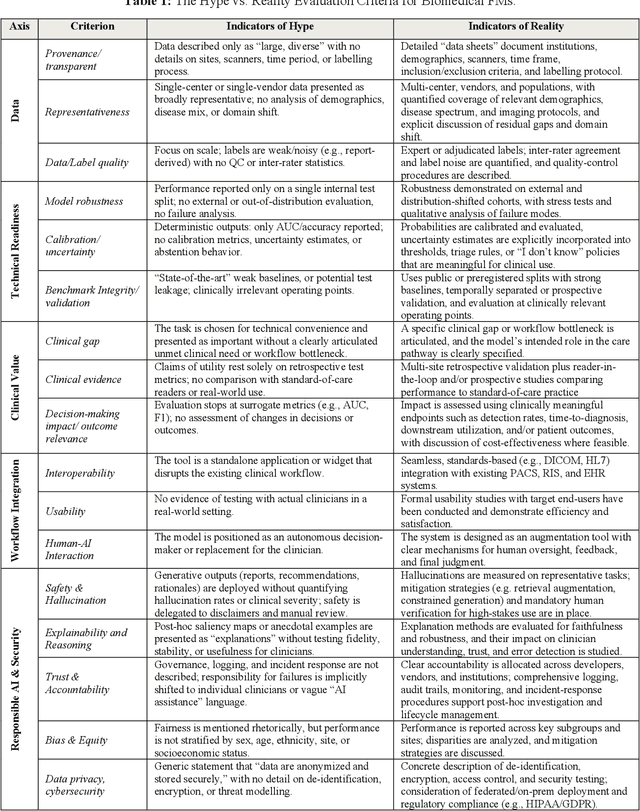
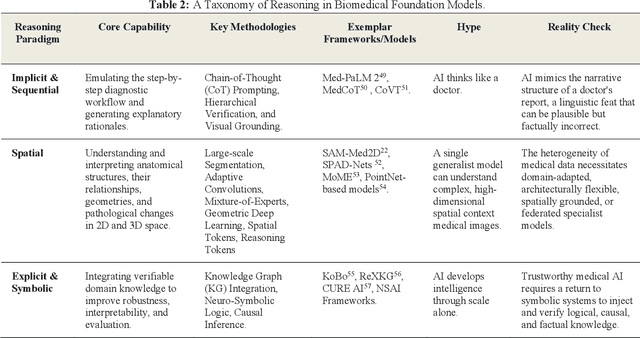
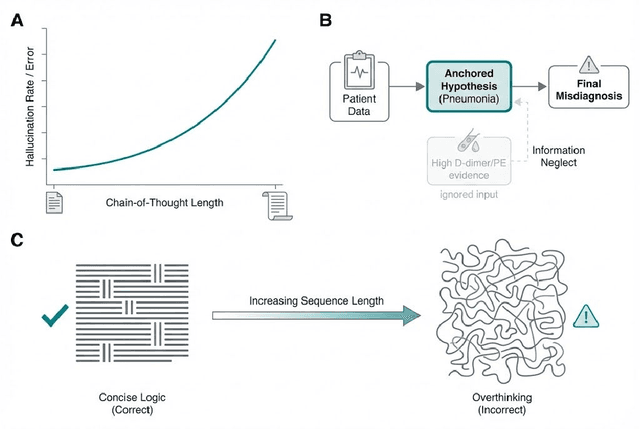
Abstract:Foundation models (FMs) are driving a prominent shift in artificial intelligence across different domains, including biomedical imaging. These models are designed to move beyond narrow pattern recognition towards emulating sophisticated clinical reasoning, understanding complex spatial relationships, and integrating multimodal data with unprecedented flexibility. However, a critical gap exists between this potential and the current reality, where the clinical evaluation and deployment of FMs are hampered by significant challenges. Herein, we critically assess the current state-of-the-art, analyzing hype by examining the core capabilities and limitations of FMs in the biomedical domain. We also provide a taxonomy of reasoning, ranging from emulated sequential logic and spatial understanding to the integration of explicit symbolic knowledge, to evaluate whether these models exhibit genuine cognition or merely mimic surface-level patterns. We argue that a critical frontier lies beyond statistical correlation, in the pursuit of causal inference, which is essential for building robust models that understand cause and effect. Furthermore, we discuss the paramount issues in deployment stemming from trustworthiness, bias, and safety, dissecting the challenges of algorithmic bias, data bias and privacy, and model hallucinations. We also draw attention to the need for more inclusive, rigorous, and clinically relevant validation frameworks to ensure their safe and ethical application. We conclude that while the vision of autonomous AI-doctors remains distant, the immediate reality is the emergence of powerful technology and assistive tools that would benefit clinical practice. The future of FMs in biomedical imaging hinges not on scale alone, but on developing hybrid, causally aware, and verifiably safe systems that augment, rather than replace, human expertise.
Cross Modality Medical Image Synthesis for Improving Liver Segmentation
Mar 02, 2025Abstract:Deep learning-based computer-aided diagnosis (CAD) of medical images requires large datasets. However, the lack of large publicly available labeled datasets limits the development of deep learning-based CAD systems. Generative Adversarial Networks (GANs), in particular, CycleGAN, can be used to generate new cross-domain images without paired training data. However, most CycleGAN-based synthesis methods lack the potential to overcome alignment and asymmetry between the input and generated data. We propose a two-stage technique for the synthesis of abdominal MRI using cross-modality translation of abdominal CT. We show that the synthetic data can help improve the performance of the liver segmentation network. We increase the number of abdominal MRI images through cross-modality image transformation of unpaired CT images using a CycleGAN inspired deformation invariant network called EssNet. Subsequently, we combine the synthetic MRI images with the original MRI images and use them to improve the accuracy of the U-Net on a liver segmentation task. We train the U-Net on real MRI images and then on real and synthetic MRI images. Consequently, by comparing both scenarios, we achieve an improvement in the performance of U-Net. In summary, the improvement achieved in the Intersection over Union (IoU) is 1.17%. The results show potential to address the data scarcity challenge in medical imaging.
Deep Learning for Surgical Instrument Recognition and Segmentation in Robotic-Assisted Surgeries: A Systematic Review
Oct 09, 2024Abstract:Applying deep learning (DL) for annotating surgical instruments in robot-assisted minimally invasive surgeries (MIS) represents a significant advancement in surgical technology. This systematic review examines 48 studies that and advanced DL methods and architectures. These sophisticated DL models have shown notable improvements in the precision and efficiency of detecting and segmenting surgical tools. The enhanced capabilities of these models support various clinical applications, including real-time intraoperative guidance, comprehensive postoperative evaluations, and objective assessments of surgical skills. By accurately identifying and segmenting surgical instruments in video data, DL models provide detailed feedback to surgeons, thereby improving surgical outcomes and reducing complication risks. Furthermore, the application of DL in surgical education is transformative. The review underscores the significant impact of DL on improving the accuracy of skill assessments and the overall quality of surgical training programs. However, implementing DL in surgical tool detection and segmentation faces challenges, such as the need for large, accurately annotated datasets to train these models effectively. The manual annotation process is labor-intensive and time-consuming, posing a significant bottleneck. Future research should focus on automating the detection and segmentation process and enhancing the robustness of DL models against environmental variations. Expanding the application of DL models across various surgical specialties will be essential to fully realize this technology's potential. Integrating DL with other emerging technologies, such as augmented reality (AR), also offers promising opportunities to further enhance the precision and efficacy of surgical procedures.
An Early Investigation into the Utility of Multimodal Large Language Models in Medical Imaging
Jun 02, 2024Abstract:Recent developments in multimodal large language models (MLLMs) have spurred significant interest in their potential applications across various medical imaging domains. On the one hand, there is a temptation to use these generative models to synthesize realistic-looking medical image data, while on the other hand, the ability to identify synthetic image data in a pool of data is also significantly important. In this study, we explore the potential of the Gemini (\textit{gemini-1.0-pro-vision-latest}) and GPT-4V (gpt-4-vision-preview) models for medical image analysis using two modalities of medical image data. Utilizing synthetic and real imaging data, both Gemini AI and GPT-4V are first used to classify real versus synthetic images, followed by an interpretation and analysis of the input images. Experimental results demonstrate that both Gemini and GPT-4 could perform some interpretation of the input images. In this specific experiment, Gemini was able to perform slightly better than the GPT-4V on the classification task. In contrast, responses associated with GPT-4V were mostly generic in nature. Our early investigation presented in this work provides insights into the potential of MLLMs to assist with the classification and interpretation of retinal fundoscopy and lung X-ray images. We also identify key limitations associated with the early investigation study on MLLMs for specialized tasks in medical image analysis.
Improving diagnosis and prognosis of lung cancer using vision transformers: A scoping review
Sep 06, 2023Abstract:Vision transformer-based methods are advancing the field of medical artificial intelligence and cancer imaging, including lung cancer applications. Recently, many researchers have developed vision transformer-based AI methods for lung cancer diagnosis and prognosis. This scoping review aims to identify the recent developments on vision transformer-based AI methods for lung cancer imaging applications. It provides key insights into how vision transformers complemented the performance of AI and deep learning methods for lung cancer. Furthermore, the review also identifies the datasets that contributed to advancing the field. Of the 314 retrieved studies, this review included 34 studies published from 2020 to 2022. The most commonly addressed task in these studies was the classification of lung cancer types, such as lung squamous cell carcinoma versus lung adenocarcinoma, and identifying benign versus malignant pulmonary nodules. Other applications included survival prediction of lung cancer patients and segmentation of lungs. The studies lacked clear strategies for clinical transformation. SWIN transformer was a popular choice of the researchers; however, many other architectures were also reported where vision transformer was combined with convolutional neural networks or UNet model. It can be concluded that vision transformer-based models are increasingly in popularity for developing AI methods for lung cancer applications. However, their computational complexity and clinical relevance are important factors to be considered for future research work. This review provides valuable insights for researchers in the field of AI and healthcare to advance the state-of-the-art in lung cancer diagnosis and prognosis. We provide an interactive dashboard on lung-cancer.onrender.com/.
Leveraging GANs for data scarcity of COVID-19: Beyond the hype
Apr 07, 2023Abstract:Artificial Intelligence (AI)-based models can help in diagnosing COVID-19 from lung CT scans and X-ray images; however, these models require large amounts of data for training and validation. Many researchers studied Generative Adversarial Networks (GANs) for producing synthetic lung CT scans and X-Ray images to improve the performance of AI-based models. It is not well explored how good GAN-based methods performed to generate reliable synthetic data. This work analyzes 43 published studies that reported GANs for synthetic data generation. Many of these studies suffered data bias, lack of reproducibility, and lack of feedback from the radiologists or other domain experts. A common issue in these studies is the unavailability of the source code, hindering reproducibility. The included studies reported rescaling of the input images to train the existing GANs architecture without providing clinical insights on how the rescaling was motivated. Finally, even though GAN-based methods have the potential for data augmentation and improving the training of AI-based models, these methods fall short in terms of their use in clinical practice. This paper highlights research hotspots in countering the data scarcity problem, identifies various issues as well as potentials, and provides recommendations to guide future research. These recommendations might be useful to improve acceptability for the GAN-based approaches for data augmentation as GANs for data augmentation are increasingly becoming popular in the AI and medical imaging research community.
Brain Tumor Synthetic Data Generation with Adaptive StyleGANs
Dec 04, 2022Abstract:Generative models have been very successful over the years and have received significant attention for synthetic data generation. As deep learning models are getting more and more complex, they require large amounts of data to perform accurately. In medical image analysis, such generative models play a crucial role as the available data is limited due to challenges related to data privacy, lack of data diversity, or uneven data distributions. In this paper, we present a method to generate brain tumor MRI images using generative adversarial networks. We have utilized StyleGAN2 with ADA methodology to generate high-quality brain MRI with tumors while using a significantly smaller amount of training data when compared to the existing approaches. We use three pre-trained models for transfer learning. Results demonstrate that the proposed method can learn the distributions of brain tumors. Furthermore, the model can generate high-quality synthetic brain MRI with a tumor that can limit the small sample size issues. The approach can addresses the limited data availability by generating realistic-looking brain MRI with tumors. The code is available at: ~\url{https://github.com/rizwanqureshi123/Brain-Tumor-Synthetic-Data}.
Spot the fake lungs: Generating Synthetic Medical Images using Neural Diffusion Models
Nov 02, 2022



Abstract:Generative models are becoming popular for the synthesis of medical images. Recently, neural diffusion models have demonstrated the potential to generate photo-realistic images of objects. However, their potential to generate medical images is not explored yet. In this work, we explore the possibilities of synthesis of medical images using neural diffusion models. First, we use a pre-trained DALLE2 model to generate lungs X-Ray and CT images from an input text prompt. Second, we train a stable diffusion model with 3165 X-Ray images and generate synthetic images. We evaluate the synthetic image data through a qualitative analysis where two independent radiologists label randomly chosen samples from the generated data as real, fake, or unsure. Results demonstrate that images generated with the diffusion model can translate characteristics that are otherwise very specific to certain medical conditions in chest X-Ray or CT images. Careful tuning of the model can be very promising. To the best of our knowledge, this is the first attempt to generate lungs X-Ray and CT images using neural diffusion models. This work aims to introduce a new dimension in artificial intelligence for medical imaging. Given that this is a new topic, the paper will serve as an introduction and motivation for the research community to explore the potential of diffusion models for medical image synthesis. We have released the synthetic images on https://www.kaggle.com/datasets/hazrat/awesomelungs.
Artificial Intelligence-Based Methods for Fusion of Electronic Health Records and Imaging Data
Oct 23, 2022Abstract:Healthcare data are inherently multimodal, including electronic health records (EHR), medical images, and multi-omics data. Combining these multimodal data sources contributes to a better understanding of human health and provides optimal personalized healthcare. Advances in artificial intelligence (AI) technologies, particularly machine learning (ML), enable the fusion of these different data modalities to provide multimodal insights. To this end, in this scoping review, we focus on synthesizing and analyzing the literature that uses AI techniques to fuse multimodal medical data for different clinical applications. More specifically, we focus on studies that only fused EHR with medical imaging data to develop various AI methods for clinical applications. We present a comprehensive analysis of the various fusion strategies, the diseases and clinical outcomes for which multimodal fusion was used, the ML algorithms used to perform multimodal fusion for each clinical application, and the available multimodal medical datasets. We followed the PRISMA-ScR guidelines. We searched Embase, PubMed, Scopus, and Google Scholar to retrieve relevant studies. We extracted data from 34 studies that fulfilled the inclusion criteria. In our analysis, a typical workflow was observed: feeding raw data, fusing different data modalities by applying conventional machine learning (ML) or deep learning (DL) algorithms, and finally, evaluating the multimodal fusion through clinical outcome predictions. Specifically, early fusion was the most used technique in most applications for multimodal learning (22 out of 34 studies). We found that multimodality fusion models outperformed traditional single-modality models for the same task. Disease diagnosis and prediction were the most common clinical outcomes (reported in 20 and 10 studies, respectively) from a clinical outcome perspective.
* Accepted in Nature Scientific Reports. 20 pages
Leveraging machine learning for less developed languages: Progress on Urdu text detection
Sep 28, 2022



Abstract:Text detection in natural scene images has applications for autonomous driving, navigation help for elderly and blind people. However, the research on Urdu text detection is usually hindered by lack of data resources. We have developed a dataset of scene images with Urdu text. We present the use of machine learning methods to perform detection of Urdu text from the scene images. We extract text regions using channel enhanced Maximally Stable Extremal Region (MSER) method. First, we classify text and noise based on their geometric properties. Next, we use a support vector machine for early discarding of non-text regions. To further remove the non-text regions, we use histogram of oriented gradients (HoG) features obtained and train a second SVM classifier. This improves the overall performance on text region detection within the scene images. To support research on Urdu text, We aim to make the data freely available for research use. We also aim to highlight the challenges and the research gap for Urdu text detection.
* Accepted at NeurIPS ML4D workshop. arXiv admin note: text overlap with arXiv:2109.08060
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge