Zubair Shah
Early Prediction of Type 2 Diabetes Using Multimodal data and Tabular Transformers
Jan 19, 2026Abstract:This study introduces a novel approach for early Type 2 Diabetes Mellitus (T2DM) risk prediction using a tabular transformer (TabTrans) architecture to analyze longitudinal patient data. By processing patients` longitudinal health records and bone-related tabular data, our model captures complex, long-range dependencies in disease progression that conventional methods often overlook. We validated our TabTrans model on a retrospective Qatar BioBank (QBB) cohort of 1,382 subjects, comprising 725 men (146 diabetic, 579 healthy) and 657 women (133 diabetic, 524 healthy). The study integrated electronic health records (EHR) with dual-energy X-ray absorptiometry (DXA) data. To address class imbalance, we employed SMOTE and SMOTE-ENN resampling techniques. The proposed model`s performance is evaluated against conventional machine learning (ML) and generative AI models, including Claude 3.5 Sonnet (Anthropic`s constitutional AI), GPT-4 (OpenAI`s generative pre-trained transformer), and Gemini Pro (Google`s multimodal language model). Our TabTrans model demonstrated superior predictive performance, achieving ROC AUC $\geq$ 79.7 % for T2DM prediction compared to both generative AI models and conventional ML approaches. Feature interpretation analysis identified key risk indicators, with visceral adipose tissue (VAT) mass and volume, ward bone mineral density (BMD) and bone mineral content (BMC), T and Z-scores, and L1-L4 scores emerging as the most important predictors associated with diabetes development in Qatari adults. These findings demonstrate the significant potential of TabTrans for analyzing complex tabular healthcare data, providing a powerful tool for proactive T2DM management and personalized clinical interventions in the Qatari population. Index Terms: tabular transformers, multimodal data, DXA data, diabetes, T2DM, feature interpretation, tabular data
Pruning as Evolution: Emergent Sparsity Through Selection Dynamics in Neural Networks
Jan 14, 2026Abstract:Neural networks are commonly trained in highly overparameterized regimes, yet empirical evidence consistently shows that many parameters become redundant during learning. Most existing pruning approaches impose sparsity through explicit intervention, such as importance-based thresholding or regularization penalties, implicitly treating pruning as a centralized decision applied to a trained model. This assumption is misaligned with the decentralized, stochastic, and path-dependent character of gradient-based training. We propose an evolutionary perspective on pruning: parameter groups (neurons, filters, heads) are modeled as populations whose influence evolves continuously under selection pressure. Under this view, pruning corresponds to population extinction: components with persistently low fitness gradually lose influence and can be removed without discrete pruning schedules and without requiring equilibrium computation. We formalize neural pruning as an evolutionary process over population masses, derive selection dynamics governing mass evolution, and connect fitness to local learning signals. We validate the framework on MNIST using a population-scaled MLP (784--512--256--10) with 768 prunable neuron populations. All dynamics reach dense baselines near 98\% test accuracy. We benchmark post-training hard pruning at target sparsity levels (35--50\%): pruning 35\% yields $\approx$95.5\% test accuracy, while pruning 50\% yields $\approx$88.3--88.6\%, depending on the dynamic. These results demonstrate that evolutionary selection produces a measurable accuracy--sparsity tradeoff without explicit pruning schedules during training.
Pruning as a Game: Equilibrium-Driven Sparsification of Neural Networks
Dec 26, 2025Abstract:Neural network pruning is widely used to reduce model size and computational cost. Yet, most existing methods treat sparsity as an externally imposed constraint, enforced through heuristic importance scores or training-time regularization. In this work, we propose a fundamentally different perspective: pruning as an equilibrium outcome of strategic interaction among model components. We model parameter groups such as weights, neurons, or filters as players in a continuous non-cooperative game, where each player selects its level of participation in the network to balance contribution against redundancy and competition. Within this formulation, sparsity emerges naturally when continued participation becomes a dominated strategy at equilibrium. We analyze the resulting game and show that dominated players collapse to zero participation under mild conditions, providing a principled explanation for pruning behavior. Building on this insight, we derive a simple equilibrium-driven pruning algorithm that jointly updates network parameters and participation variables without relying on explicit importance scores. This work focuses on establishing a principled formulation and empirical validation of pruning as an equilibrium phenomenon, rather than exhaustive architectural or large-scale benchmarking. Experiments on standard benchmarks demonstrate that the proposed approach achieves competitive sparsity-accuracy trade-offs while offering an interpretable, theory-grounded alternative to existing pruning methods.
Federated Learning of Low-Rank One-Shot Image Detection Models in Edge Devices with Scalable Accuracy and Compute Complexity
Apr 23, 2025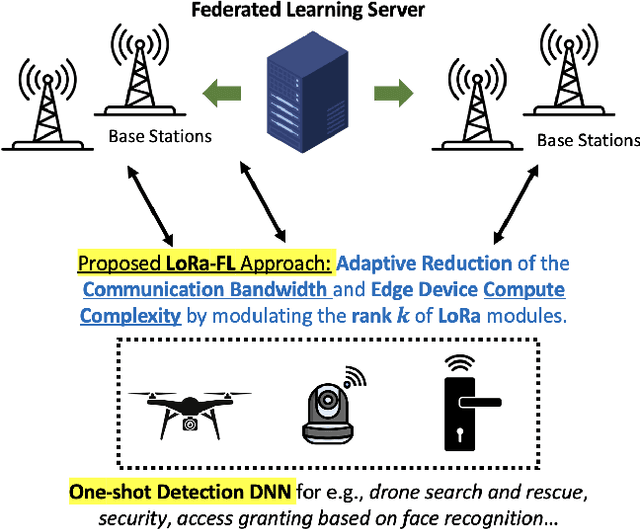
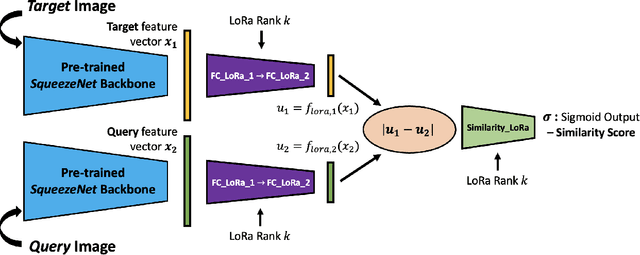
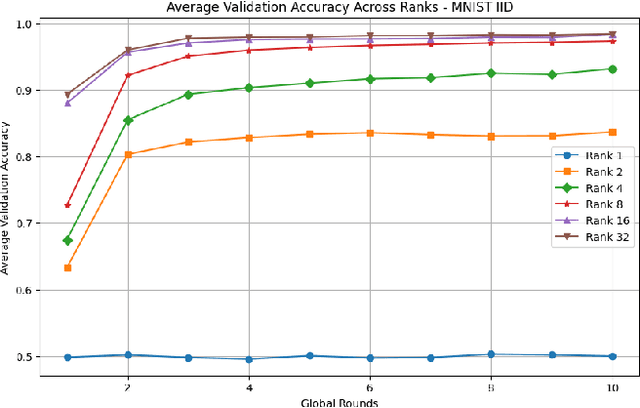
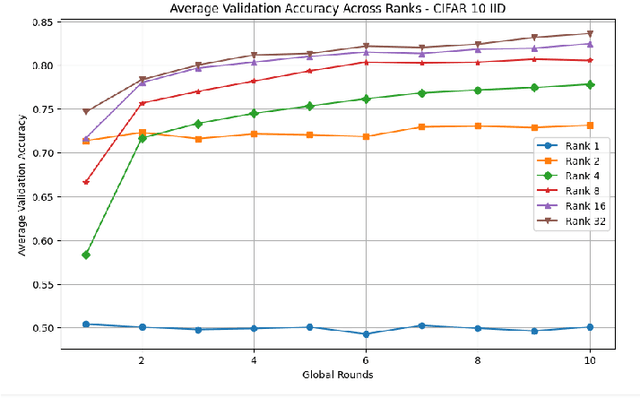
Abstract:This paper introduces a novel federated learning framework termed LoRa-FL designed for training low-rank one-shot image detection models deployed on edge devices. By incorporating low-rank adaptation techniques into one-shot detection architectures, our method significantly reduces both computational and communication overhead while maintaining scalable accuracy. The proposed framework leverages federated learning to collaboratively train lightweight image recognition models, enabling rapid adaptation and efficient deployment across heterogeneous, resource-constrained devices. Experimental evaluations on the MNIST and CIFAR10 benchmark datasets, both in an independent-and-identically-distributed (IID) and non-IID setting, demonstrate that our approach achieves competitive detection performance while significantly reducing communication bandwidth and compute complexity. This makes it a promising solution for adaptively reducing the communication and compute power overheads, while not sacrificing model accuracy.
Integrating Non-Linear Radon Transformation for Diabetic Retinopathy Grading
Apr 22, 2025Abstract:Diabetic retinopathy is a serious ocular complication that poses a significant threat to patients' vision and overall health. Early detection and accurate grading are essential to prevent vision loss. Current automatic grading methods rely heavily on deep learning applied to retinal fundus images, but the complex, irregular patterns of lesions in these images, which vary in shape and distribution, make it difficult to capture subtle changes. This study introduces RadFuse, a multi-representation deep learning framework that integrates non-linear RadEx-transformed sinogram images with traditional fundus images to enhance diabetic retinopathy detection and grading. Our RadEx transformation, an optimized non-linear extension of the Radon transform, generates sinogram representations to capture complex retinal lesion patterns. By leveraging both spatial and transformed domain information, RadFuse enriches the feature set available to deep learning models, improving the differentiation of severity levels. We conducted extensive experiments on two benchmark datasets, APTOS-2019 and DDR, using three convolutional neural networks (CNNs): ResNeXt-50, MobileNetV2, and VGG19. RadFuse showed significant improvements over fundus-image-only models across all three CNN architectures and outperformed state-of-the-art methods on both datasets. For severity grading across five stages, RadFuse achieved a quadratic weighted kappa of 93.24%, an accuracy of 87.07%, and an F1-score of 87.17%. In binary classification between healthy and diabetic retinopathy cases, the method reached an accuracy of 99.09%, precision of 98.58%, and recall of 99.6%, surpassing previously established models. These results demonstrate RadFuse's capacity to capture complex non-linear features, advancing diabetic retinopathy classification and promoting the integration of advanced mathematical transforms in medical image analysis.
Improving Early Prediction of Type 2 Diabetes Mellitus with ECG-DiaNet: A Multimodal Neural Network Leveraging Electrocardiogram and Clinical Risk Factors
Apr 05, 2025Abstract:Type 2 Diabetes Mellitus (T2DM) remains a global health challenge, underscoring the need for early and accurate risk prediction. This study presents ECG-DiaNet, a multimodal deep learning model that integrates electrocardiogram (ECG) features with clinical risk factors (CRFs) to enhance T2DM onset prediction. Using data from Qatar Biobank (QBB), we trained and validated models on a development cohort (n=2043) and evaluated performance on a longitudinal test set (n=395) with five-year follow-up. ECG-DiaNet outperformed unimodal ECG-only and CRF-only models, achieving a higher AUROC (0.845 vs 0.8217) than the CRF-only model, with statistical significance (DeLong p<0.001). Reclassification metrics further confirmed improvements: Net Reclassification Improvement (NRI=0.0153) and Integrated Discrimination Improvement (IDI=0.0482). Risk stratification into low-, medium-, and high-risk groups showed ECG-DiaNet achieved superior positive predictive value (PPV) in high-risk individuals. The model's reliance on non-invasive and widely available ECG signals supports its feasibility in clinical and community health settings. By combining cardiac electrophysiology and systemic risk profiles, ECG-DiaNet addresses the multifactorial nature of T2DM and supports precision prevention. These findings highlight the value of multimodal AI in advancing early detection and prevention strategies for T2DM, particularly in underrepresented Middle Eastern populations.
Cross Modality Medical Image Synthesis for Improving Liver Segmentation
Mar 02, 2025Abstract:Deep learning-based computer-aided diagnosis (CAD) of medical images requires large datasets. However, the lack of large publicly available labeled datasets limits the development of deep learning-based CAD systems. Generative Adversarial Networks (GANs), in particular, CycleGAN, can be used to generate new cross-domain images without paired training data. However, most CycleGAN-based synthesis methods lack the potential to overcome alignment and asymmetry between the input and generated data. We propose a two-stage technique for the synthesis of abdominal MRI using cross-modality translation of abdominal CT. We show that the synthetic data can help improve the performance of the liver segmentation network. We increase the number of abdominal MRI images through cross-modality image transformation of unpaired CT images using a CycleGAN inspired deformation invariant network called EssNet. Subsequently, we combine the synthetic MRI images with the original MRI images and use them to improve the accuracy of the U-Net on a liver segmentation task. We train the U-Net on real MRI images and then on real and synthetic MRI images. Consequently, by comparing both scenarios, we achieve an improvement in the performance of U-Net. In summary, the improvement achieved in the Intersection over Union (IoU) is 1.17%. The results show potential to address the data scarcity challenge in medical imaging.
Deep Learning for Surgical Instrument Recognition and Segmentation in Robotic-Assisted Surgeries: A Systematic Review
Oct 09, 2024Abstract:Applying deep learning (DL) for annotating surgical instruments in robot-assisted minimally invasive surgeries (MIS) represents a significant advancement in surgical technology. This systematic review examines 48 studies that and advanced DL methods and architectures. These sophisticated DL models have shown notable improvements in the precision and efficiency of detecting and segmenting surgical tools. The enhanced capabilities of these models support various clinical applications, including real-time intraoperative guidance, comprehensive postoperative evaluations, and objective assessments of surgical skills. By accurately identifying and segmenting surgical instruments in video data, DL models provide detailed feedback to surgeons, thereby improving surgical outcomes and reducing complication risks. Furthermore, the application of DL in surgical education is transformative. The review underscores the significant impact of DL on improving the accuracy of skill assessments and the overall quality of surgical training programs. However, implementing DL in surgical tool detection and segmentation faces challenges, such as the need for large, accurately annotated datasets to train these models effectively. The manual annotation process is labor-intensive and time-consuming, posing a significant bottleneck. Future research should focus on automating the detection and segmentation process and enhancing the robustness of DL models against environmental variations. Expanding the application of DL models across various surgical specialties will be essential to fully realize this technology's potential. Integrating DL with other emerging technologies, such as augmented reality (AR), also offers promising opportunities to further enhance the precision and efficacy of surgical procedures.
MemeMind at ArAIEval Shared Task: Spotting Persuasive Spans in Arabic Text with Persuasion Techniques Identification
Aug 08, 2024
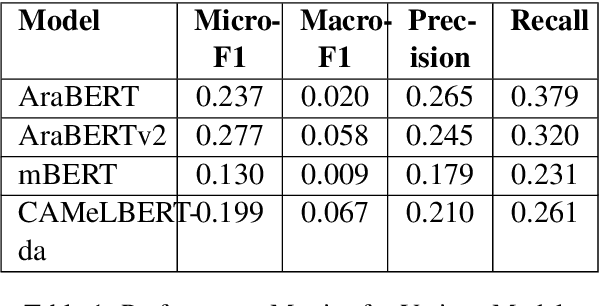
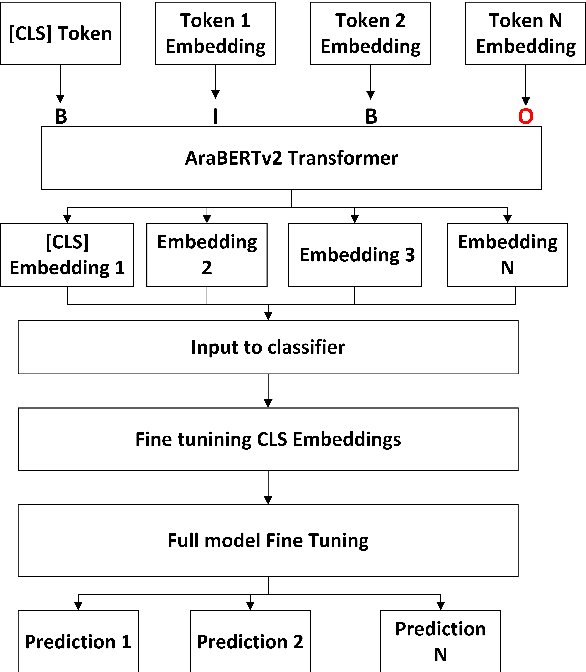

Abstract:This paper focuses on detecting propagandistic spans and persuasion techniques in Arabic text from tweets and news paragraphs. Each entry in the dataset contains a text sample and corresponding labels that indicate the start and end positions of propaganda techniques within the text. Tokens falling within a labeled span were assigned "B" (Begin) or "I" (Inside), "O", corresponding to the specific propaganda technique. Using attention masks, we created uniform lengths for each span and assigned BIO tags to each token based on the provided labels. Then, we used AraBERT-base pre-trained model for Arabic text tokenization and embeddings with a token classification layer to identify propaganda techniques. Our training process involves a two-phase fine-tuning approach. First, we train only the classification layer for a few epochs, followed by full model fine-tuning, updating all parameters. This methodology allows the model to adapt to the specific characteristics of the propaganda detection task while leveraging the knowledge captured by the pre-trained AraBERT model. Our approach achieved an F1 score of 0.2774, securing the 3rd position in the leaderboard of Task 1.
An Early Investigation into the Utility of Multimodal Large Language Models in Medical Imaging
Jun 02, 2024Abstract:Recent developments in multimodal large language models (MLLMs) have spurred significant interest in their potential applications across various medical imaging domains. On the one hand, there is a temptation to use these generative models to synthesize realistic-looking medical image data, while on the other hand, the ability to identify synthetic image data in a pool of data is also significantly important. In this study, we explore the potential of the Gemini (\textit{gemini-1.0-pro-vision-latest}) and GPT-4V (gpt-4-vision-preview) models for medical image analysis using two modalities of medical image data. Utilizing synthetic and real imaging data, both Gemini AI and GPT-4V are first used to classify real versus synthetic images, followed by an interpretation and analysis of the input images. Experimental results demonstrate that both Gemini and GPT-4 could perform some interpretation of the input images. In this specific experiment, Gemini was able to perform slightly better than the GPT-4V on the classification task. In contrast, responses associated with GPT-4V were mostly generic in nature. Our early investigation presented in this work provides insights into the potential of MLLMs to assist with the classification and interpretation of retinal fundoscopy and lung X-ray images. We also identify key limitations associated with the early investigation study on MLLMs for specialized tasks in medical image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge