Mingxing Li
Searth Transformer: A Transformer Architecture Incorporating Earth's Geospheric Physical Priors for Global Mid-Range Weather Forecasting
Jan 14, 2026Abstract:Accurate global medium-range weather forecasting is fundamental to Earth system science. Most existing Transformer-based forecasting models adopt vision-centric architectures that neglect the Earth's spherical geometry and zonal periodicity. In addition, conventional autoregressive training is computationally expensive and limits forecast horizons due to error accumulation. To address these challenges, we propose the Shifted Earth Transformer (Searth Transformer), a physics-informed architecture that incorporates zonal periodicity and meridional boundaries into window-based self-attention for physically consistent global information exchange. We further introduce a Relay Autoregressive (RAR) fine-tuning strategy that enables learning long-range atmospheric evolution under constrained memory and computational budgets. Based on these methods, we develop YanTian, a global medium-range weather forecasting model. YanTian achieves higher accuracy than the high-resolution forecast of the European Centre for Medium-Range Weather Forecasts and performs competitively with state-of-the-art AI models at one-degree resolution, while requiring roughly 200 times lower computational cost than standard autoregressive fine-tuning. Furthermore, YanTian attains a longer skillful forecast lead time for Z500 (10.3 days) than HRES (9 days). Beyond weather forecasting, this work establishes a robust algorithmic foundation for predictive modeling of complex global-scale geophysical circulation systems, offering new pathways for Earth system science.
From Editor to Dense Geometry Estimator
Sep 04, 2025
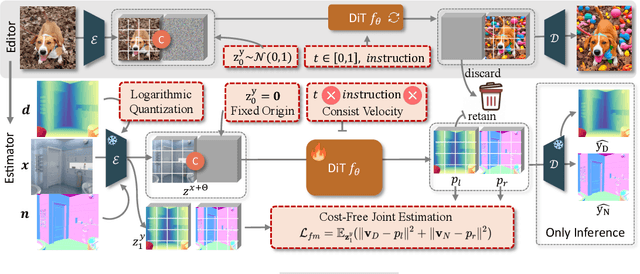


Abstract:Leveraging visual priors from pre-trained text-to-image (T2I) generative models has shown success in dense prediction. However, dense prediction is inherently an image-to-image task, suggesting that image editing models, rather than T2I generative models, may be a more suitable foundation for fine-tuning. Motivated by this, we conduct a systematic analysis of the fine-tuning behaviors of both editors and generators for dense geometry estimation. Our findings show that editing models possess inherent structural priors, which enable them to converge more stably by ``refining" their innate features, and ultimately achieve higher performance than their generative counterparts. Based on these findings, we introduce \textbf{FE2E}, a framework that pioneeringly adapts an advanced editing model based on Diffusion Transformer (DiT) architecture for dense geometry prediction. Specifically, to tailor the editor for this deterministic task, we reformulate the editor's original flow matching loss into the ``consistent velocity" training objective. And we use logarithmic quantization to resolve the precision conflict between the editor's native BFloat16 format and the high precision demand of our tasks. Additionally, we leverage the DiT's global attention for a cost-free joint estimation of depth and normals in a single forward pass, enabling their supervisory signals to mutually enhance each other. Without scaling up the training data, FE2E achieves impressive performance improvements in zero-shot monocular depth and normal estimation across multiple datasets. Notably, it achieves over 35\% performance gains on the ETH3D dataset and outperforms the DepthAnything series, which is trained on 100$\times$ data. The project page can be accessed \href{https://amap-ml.github.io/FE2E/}{here}.
UniVG-R1: Reasoning Guided Universal Visual Grounding with Reinforcement Learning
May 20, 2025Abstract:Traditional visual grounding methods primarily focus on single-image scenarios with simple textual references. However, extending these methods to real-world scenarios that involve implicit and complex instructions, particularly in conjunction with multiple images, poses significant challenges, which is mainly due to the lack of advanced reasoning ability across diverse multi-modal contexts. In this work, we aim to address the more practical universal grounding task, and propose UniVG-R1, a reasoning guided multimodal large language model (MLLM) for universal visual grounding, which enhances reasoning capabilities through reinforcement learning (RL) combined with cold-start data. Specifically, we first construct a high-quality Chain-of-Thought (CoT) grounding dataset, annotated with detailed reasoning chains, to guide the model towards correct reasoning paths via supervised fine-tuning. Subsequently, we perform rule-based reinforcement learning to encourage the model to identify correct reasoning chains, thereby incentivizing its reasoning capabilities. In addition, we identify a difficulty bias arising from the prevalence of easy samples as RL training progresses, and we propose a difficulty-aware weight adjustment strategy to further strengthen the performance. Experimental results demonstrate the effectiveness of UniVG-R1, which achieves state-of-the-art performance on MIG-Bench with a 9.1% improvement over the previous method. Furthermore, our model exhibits strong generalizability, achieving an average improvement of 23.4% in zero-shot performance across four image and video reasoning grounding benchmarks. The project page can be accessed at https://amap-ml.github.io/UniVG-R1-page/.
FLUX-Text: A Simple and Advanced Diffusion Transformer Baseline for Scene Text Editing
May 06, 2025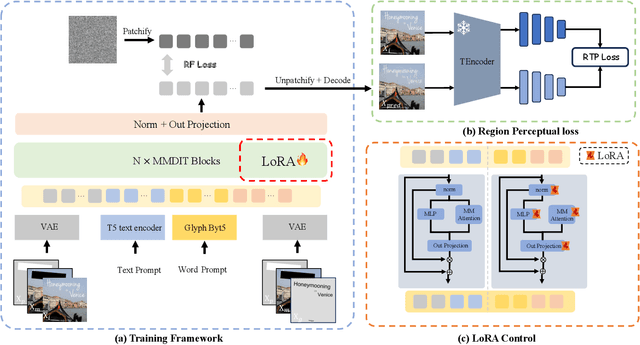
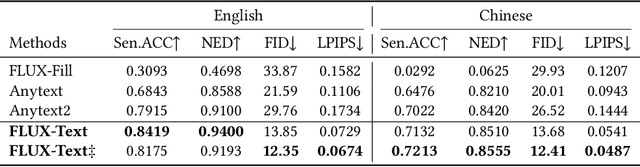
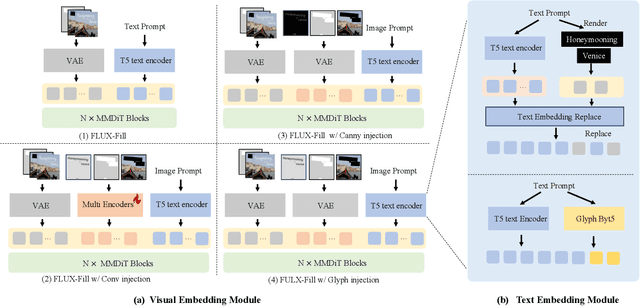
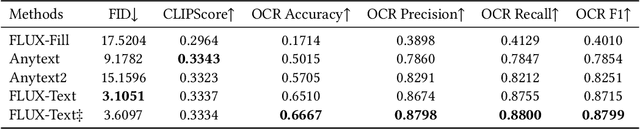
Abstract:The task of scene text editing is to modify or add texts on images while maintaining the fidelity of newly generated text and visual coherence with the background. Recent works based on latent diffusion models (LDM) show improved text editing results, yet still face challenges and often generate inaccurate or unrecognizable characters, especially for non-Latin ones (\eg, Chinese), which have complex glyph structures. To address these issues, we present FLUX-Text, a simple and advanced multilingual scene text editing framework based on FLUX-Fill. Specifically, we carefully investigate glyph conditioning, considering both visual and textual modalities. To retain the original generative capabilities of FLUX-Fill while enhancing its understanding and generation of glyphs, we propose lightweight glyph and text embedding modules. Owning to the lightweight design, FLUX-Text is trained only with $100K$ training examples compared to current popular methods trained with 2.9M ones. With no bells and whistles, our method achieves state-of-the-art performance on text editing tasks. Qualitative and quantitative experiments on the public datasets demonstrate that our method surpasses previous works in text fidelity.
Next Token Is Enough: Realistic Image Quality and Aesthetic Scoring with Multimodal Large Language Model
Mar 08, 2025Abstract:The rapid expansion of mobile internet has resulted in a substantial increase in user-generated content (UGC) images, thereby making the thorough assessment of UGC images both urgent and essential. Recently, multimodal large language models (MLLMs) have shown great potential in image quality assessment (IQA) and image aesthetic assessment (IAA). Despite this progress, effectively scoring the quality and aesthetics of UGC images still faces two main challenges: 1) A single score is inadequate to capture the hierarchical human perception. 2) How to use MLLMs to output numerical scores, such as mean opinion scores (MOS), remains an open question. To address these challenges, we introduce a novel dataset, named Realistic image Quality and Aesthetic (RealQA), including 14,715 UGC images, each of which is annoted with 10 fine-grained attributes. These attributes span three levels: low level (e.g., image clarity), middle level (e.g., subject integrity) and high level (e.g., composition). Besides, we conduct a series of in-depth and comprehensive investigations into how to effectively predict numerical scores using MLLMs. Surprisingly, by predicting just two extra significant digits, the next token paradigm can achieve SOTA performance. Furthermore, with the help of chain of thought (CoT) combined with the learnt fine-grained attributes, the proposed method can outperform SOTA methods on five public datasets for IQA and IAA with superior interpretability and show strong zero-shot generalization for video quality assessment (VQA). The code and dataset will be released.
Why is the winner the best?
Mar 30, 2023
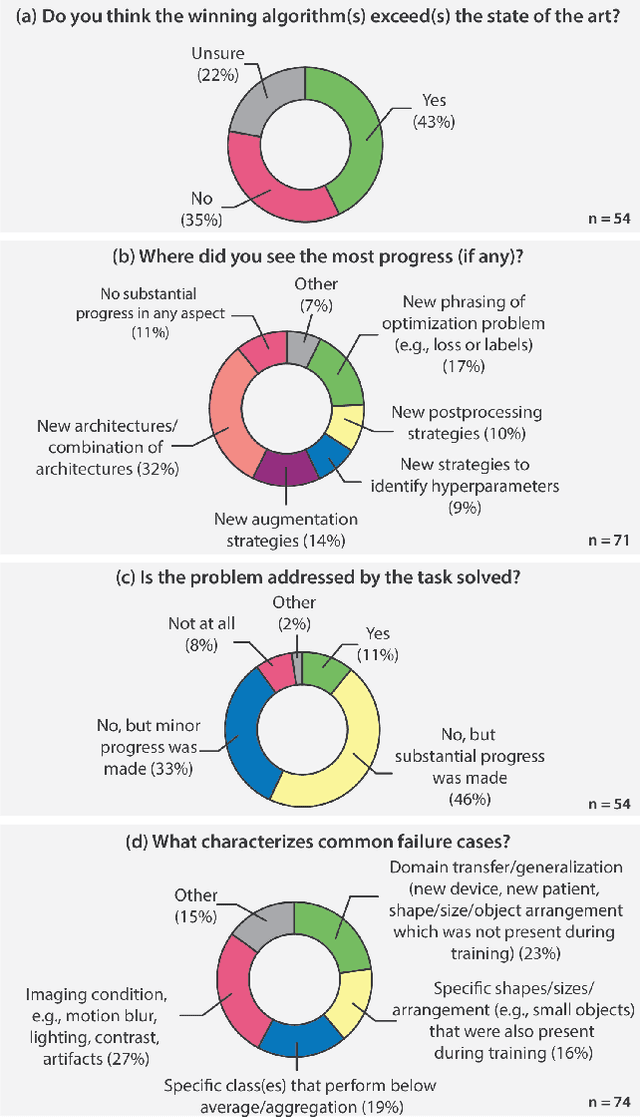
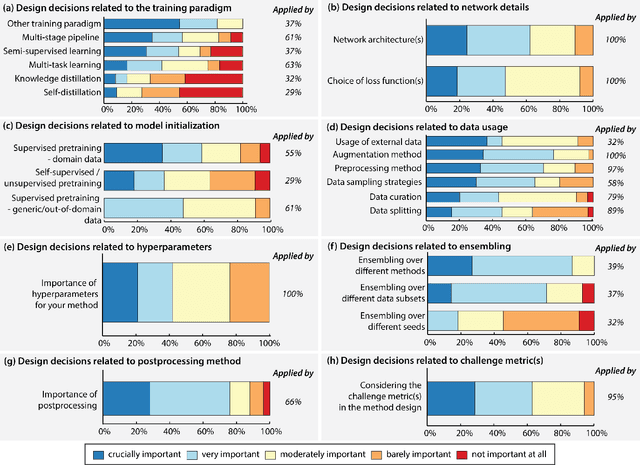
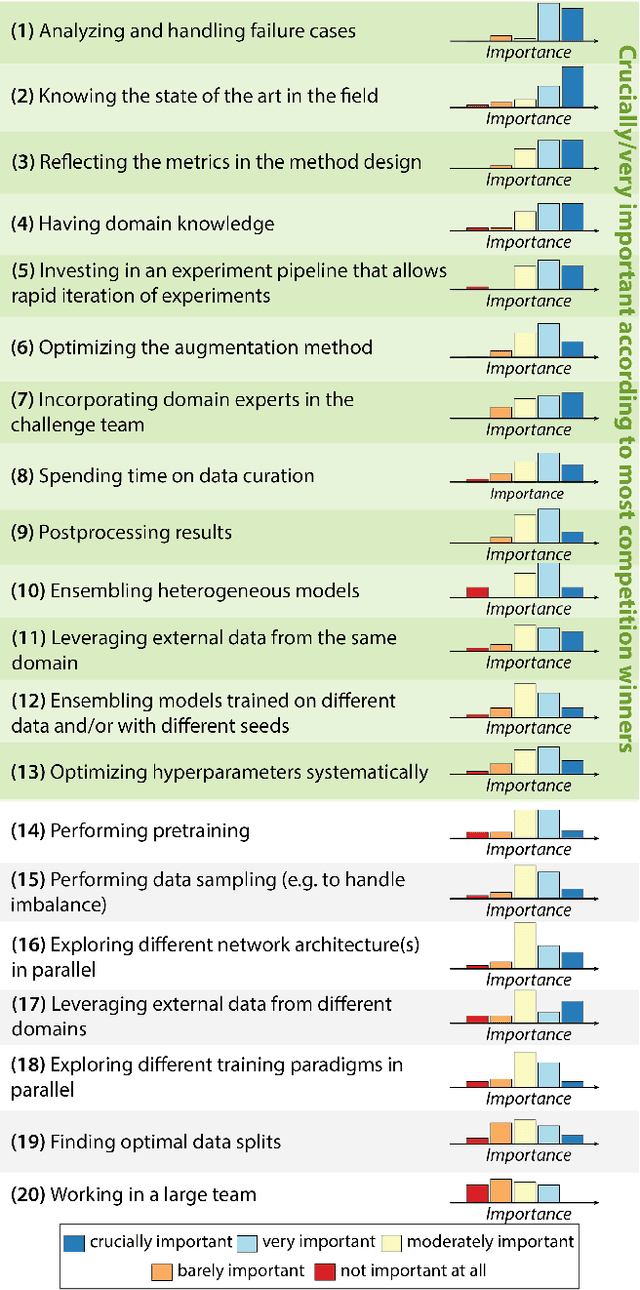
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Recurrent Dynamic Embedding for Video Object Segmentation
May 08, 2022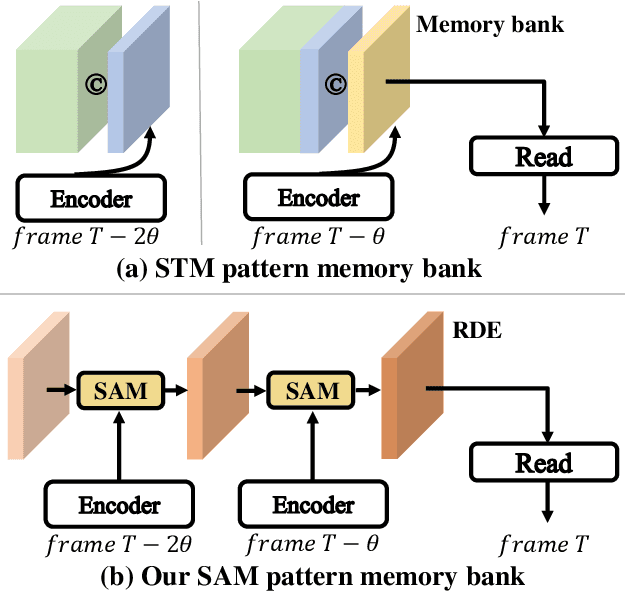
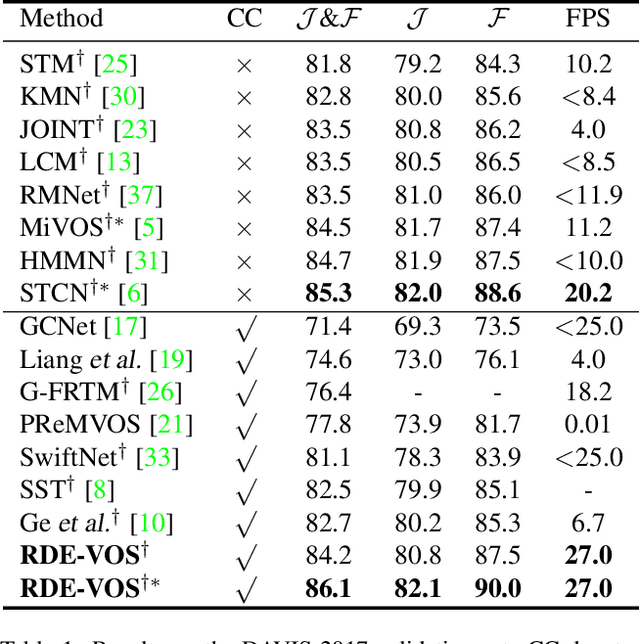
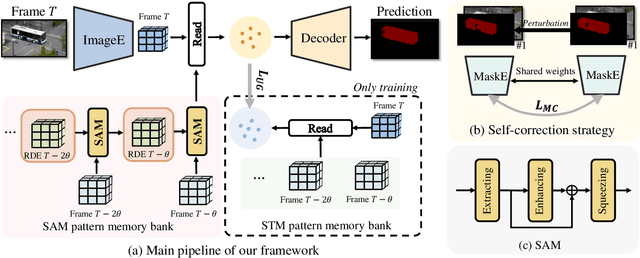
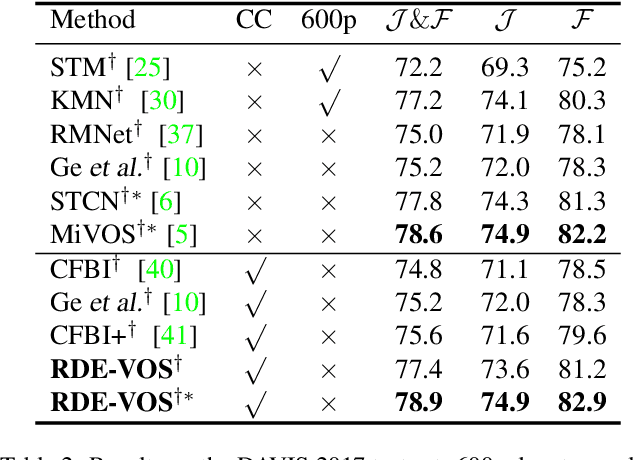
Abstract:Space-time memory (STM) based video object segmentation (VOS) networks usually keep increasing memory bank every several frames, which shows excellent performance. However, 1) the hardware cannot withstand the ever-increasing memory requirements as the video length increases. 2) Storing lots of information inevitably introduces lots of noise, which is not conducive to reading the most important information from the memory bank. In this paper, we propose a Recurrent Dynamic Embedding (RDE) to build a memory bank of constant size. Specifically, we explicitly generate and update RDE by the proposed Spatio-temporal Aggregation Module (SAM), which exploits the cue of historical information. To avoid error accumulation owing to the recurrent usage of SAM, we propose an unbiased guidance loss during the training stage, which makes SAM more robust in long videos. Moreover, the predicted masks in the memory bank are inaccurate due to the inaccurate network inference, which affects the segmentation of the query frame. To address this problem, we design a novel self-correction strategy so that the network can repair the embeddings of masks with different qualities in the memory bank. Extensive experiments show our method achieves the best tradeoff between performance and speed. Code is available at https://github.com/Limingxing00/RDE-VOS-CVPR2022.
Retinal Vessel Segmentation with Pixel-wise Adaptive Filters
Feb 03, 2022


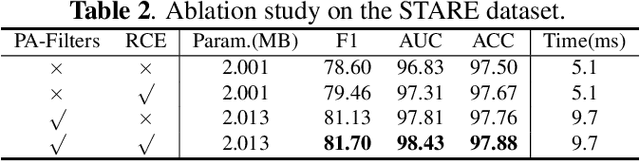
Abstract:Accurate retinal vessel segmentation is challenging because of the complex texture of retinal vessels and low imaging contrast. Previous methods generally refine segmentation results by cascading multiple deep networks, which are time-consuming and inefficient. In this paper, we propose two novel methods to address these challenges. First, we devise a light-weight module, named multi-scale residual similarity gathering (MRSG), to generate pixel-wise adaptive filters (PA-Filters). Different from cascading multiple deep networks, only one PA-Filter layer can improve the segmentation results. Second, we introduce a response cue erasing (RCE) strategy to enhance the segmentation accuracy. Experimental results on the DRIVE, CHASE_DB1, and STARE datasets demonstrate that our proposed method outperforms state-of-the-art methods while maintaining a compact structure. Code is available at https://github.com/Limingxing00/Retinal-Vessel-Segmentation-ISBI20222.
Advanced Deep Networks for 3D Mitochondria Instance Segmentation
Apr 16, 2021
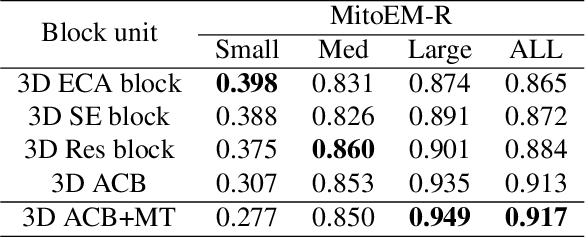


Abstract:Mitochondria instance segmentation from electron microscopy (EM) images has seen notable progress since the introduction of deep learning methods. In this paper, we propose two advanced deep networks, named Res-UNet-R and Res-UNet-H, for 3D mitochondria instance segmentation from Rat and Human samples. Specifically, we design a simple yet effective anisotropic convolution block and deploy a multi-scale training strategy, which together boost the segmentation performance. Moreover, we enhance the generalizability of the trained models on the test set by adding a denoising operation as pre-processing. In the Large-scale 3D Mitochondria Instance Segmentation Challenge, our team ranks the 1st on the leaderboard at the end of the testing phase. Code is available at https://github.com/Limingxing00/MitoEM2021-Challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge