Lingyan Zhang
Robust Simultaneous Multislice MRI Reconstruction Using Deep Generative Priors
Jul 31, 2024



Abstract:Simultaneous multislice (SMS) imaging is a powerful technique for accelerating magnetic resonance imaging (MRI) acquisitions. However, SMS reconstruction remains challenging due to the complex signal interactions between and within the excited slices. This study presents a robust SMS MRI reconstruction method using deep generative priors. Starting from Gaussian noise, we leverage denoising diffusion probabilistic models (DDPM) to gradually recover the individual slices through reverse diffusion iterations while imposing data consistency from the measured k-space under readout concatenation framework. The posterior sampling procedure is designed such that the DDPM training can be performed on single-slice images without special adjustments for SMS tasks. Additionally, our method integrates a low-frequency enhancement (LFE) module to address a practical issue that SMS-accelerated fast spin echo (FSE) and echo-planar imaging (EPI) sequences cannot easily embed autocalibration signals. Extensive experiments demonstrate that our approach consistently outperforms existing methods and generalizes well to unseen datasets. The code is available at https://github.com/Solor-pikachu/ROGER after the review process.
Noise Level Adaptive Diffusion Model for Robust Reconstruction of Accelerated MRI
Mar 08, 2024Abstract:In general, diffusion model-based MRI reconstruction methods incrementally remove artificially added noise while imposing data consistency to reconstruct the underlying images. However, real-world MRI acquisitions already contain inherent noise due to thermal fluctuations. This phenomenon is particularly notable when using ultra-fast, high-resolution imaging sequences for advanced research, or using low-field systems favored by low- and middle-income countries. These common scenarios can lead to sub-optimal performance or complete failure of existing diffusion model-based reconstruction techniques. Specifically, as the artificially added noise is gradually removed, the inherent MRI noise becomes increasingly pronounced, making the actual noise level inconsistent with the predefined denoising schedule and consequently inaccurate image reconstruction. To tackle this problem, we propose a posterior sampling strategy with a novel NoIse Level Adaptive Data Consistency (Nila-DC) operation. Extensive experiments are conducted on two public datasets and an in-house clinical dataset with field strength ranging from 0.3T to 3T, showing that our method surpasses the state-of-the-art MRI reconstruction methods, and is highly robust against various noise levels. The code will be released after review.
Automatic Radio Map Adaptation for Robust Localization with Dynamic Adversarial Learning
Feb 19, 2024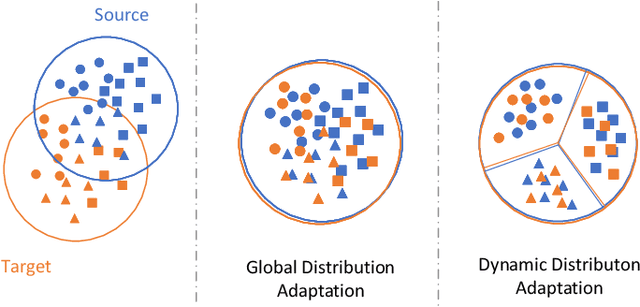
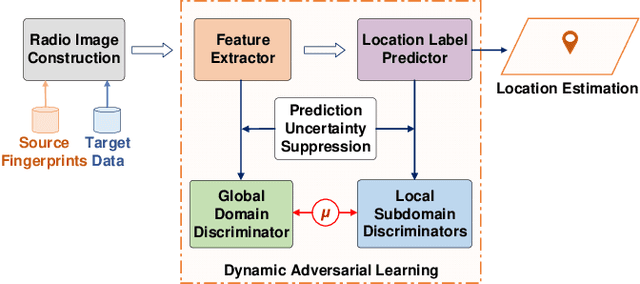
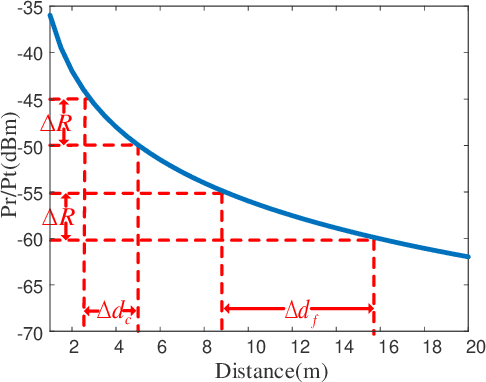
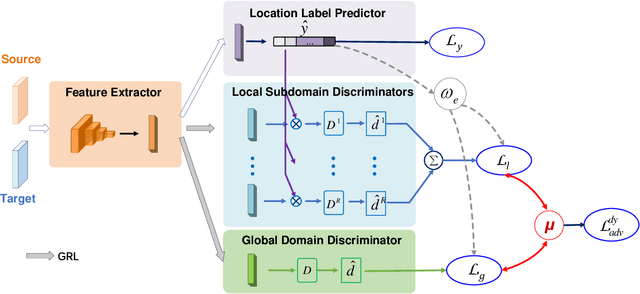
Abstract:Wireless fingerprint-based localization has become one of the most promising technologies for ubiquitous location-aware computing and intelligent location-based services. However, due to RF vulnerability to environmental dynamics over time, continuous radio map updates are time-consuming and infeasible, resulting in severe accuracy degradation. To address this issue, we propose a novel approach of robust localization with dynamic adversarial learning, known as DadLoc which realizes automatic radio map adaptation by incorporating multiple robust factors underlying RF fingerprints to learn the evolving feature representation with the complicated environmental dynamics. DadLoc performs a finer-grained distribution adaptation with the developed dynamic adversarial adaptation network and quantifies the contributions of both global and local distribution adaptation in a dynamics-adaptive manner. Furthermore, we adopt the strategy of prediction uncertainty suppression to conduct source-supervised training, target-unsupervised training, and source-target dynamic adversarial adaptation which can trade off the environment adaptability and the location discriminability of the learned deep representation for safe and effective feature transfer across different environments. With extensive experimental results, the satisfactory accuracy over other comparative schemes demonstrates that the proposed DanLoc can facilitate fingerprint-based localization for wide deployments.
ICHPro: Intracerebral Hemorrhage Prognosis Classification Via Joint-attention Fusion-based 3d Cross-modal Network
Feb 17, 2024
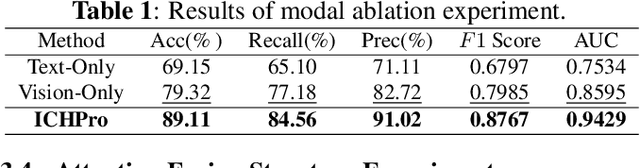

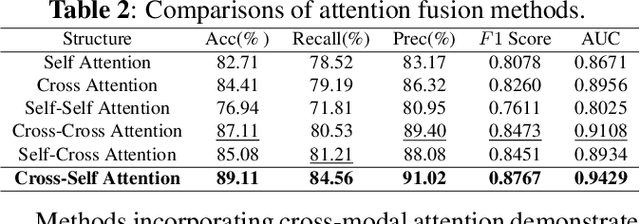
Abstract:Intracerebral Hemorrhage (ICH) is the deadliest subtype of stroke, necessitating timely and accurate prognostic evaluation to reduce mortality and disability. However, the multi-factorial nature and complexity of ICH make methods based solely on computed tomography (CT) image features inadequate. Despite the capacity of cross-modal networks to fuse additional information, the effective combination of different modal features remains a significant challenge. In this study, we propose a joint-attention fusion-based 3D cross-modal network termed ICHPro that simulates the ICH prognosis interpretation process utilized by neurosurgeons. ICHPro includes a joint-attention fusion module to fuse features from CT images with demographic and clinical textual data. To enhance the representation of cross-modal features, we introduce a joint loss function. ICHPro facilitates the extraction of richer cross-modal features, thereby improving classification performance. Upon testing our method using a five-fold cross-validation, we achieved an accuracy of 89.11%, an F1 score of 0.8767, and an AUC value of 0.9429. These results outperform those obtained from other advanced methods based on the test dataset, thereby demonstrating the superior efficacy of ICHPro. The code is available at our Github: https://github.com/YU-deep/ICH.
GCS-ICHNet: Assessment of Intracerebral Hemorrhage Prognosis using Self-Attention with Domain Knowledge Integration
Nov 08, 2023



Abstract:Intracerebral Hemorrhage (ICH) is a severe condition resulting from damaged brain blood vessel ruptures, often leading to complications and fatalities. Timely and accurate prognosis and management are essential due to its high mortality rate. However, conventional methods heavily rely on subjective clinician expertise, which can lead to inaccurate diagnoses and delays in treatment. Artificial intelligence (AI) models have been explored to assist clinicians, but many prior studies focused on model modification without considering domain knowledge. This paper introduces a novel deep learning algorithm, GCS-ICHNet, which integrates multimodal brain CT image data and the Glasgow Coma Scale (GCS) score to improve ICH prognosis. The algorithm utilizes a transformer-based fusion module for assessment. GCS-ICHNet demonstrates high sensitivity 81.03% and specificity 91.59%, outperforming average clinicians and other state-of-the-art methods.
Toward Unpaired Multi-modal Medical Image Segmentation via Learning Structured Semantic Consistency
Jun 21, 2022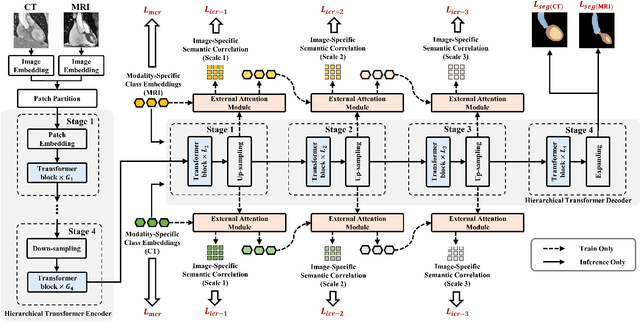
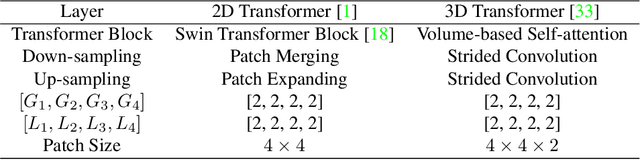
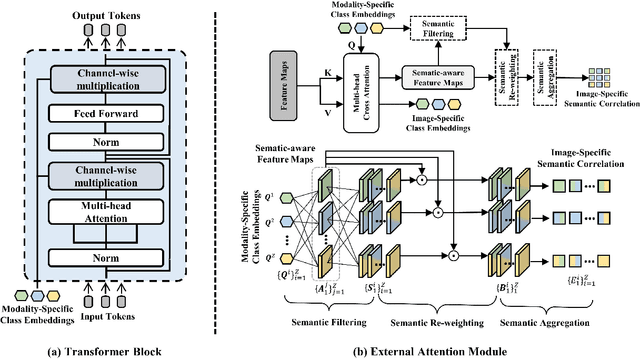
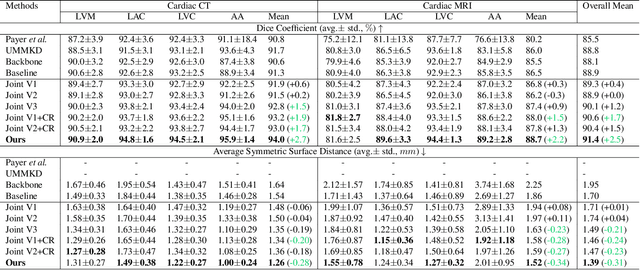
Abstract:Integrating multi-modal data to improve medical image analysis has received great attention recently. However, due to the modal discrepancy, how to use a single model to process the data from multiple modalities is still an open issue. In this paper, we propose a novel scheme to achieve better pixel-level segmentation for unpaired multi-modal medical images. Different from previous methods which adopted both modality-specific and modality-shared modules to accommodate the appearance variance of different modalities while extracting the common semantic information, our method is based on a single Transformer with a carefully designed External Attention Module (EAM) to learn the structured semantic consistency (i.e. semantic class representations and their correlations) between modalities in the training phase. In practice, the above-mentioned structured semantic consistency across modalities can be progressively achieved by implementing the consistency regularization at the modality-level and image-level respectively. The proposed EAMs are adopted to learn the semantic consistency for different scale representations and can be discarded once the model is optimized. Therefore, during the testing phase, we only need to maintain one Transformer for all modal predictions, which nicely balances the model's ease of use and simplicity. To demonstrate the effectiveness of the proposed method, we conduct the experiments on two medical image segmentation scenarios: (1) cardiac structure segmentation, and (2) abdominal multi-organ segmentation. Extensive results show that the proposed method outperforms the state-of-the-art methods by a wide margin, and even achieves competitive performance with extremely limited training samples (e.g., 1 or 3 annotated CT or MRI images) for one specific modality.
AMOS: A Large-Scale Abdominal Multi-Organ Benchmark for Versatile Medical Image Segmentation
Jun 16, 2022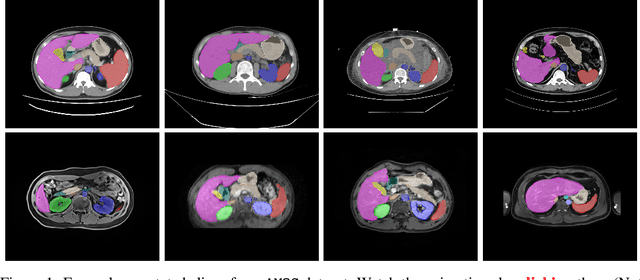
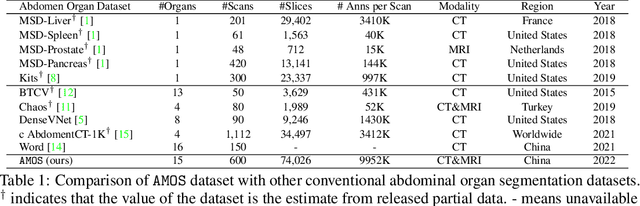
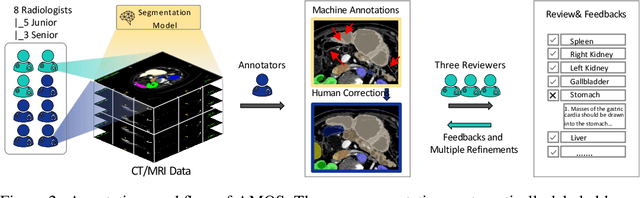
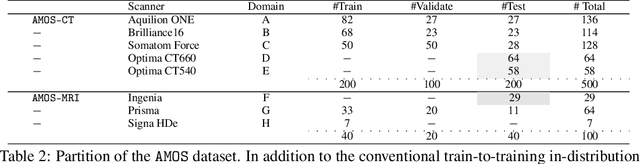
Abstract:Despite the considerable progress in automatic abdominal multi-organ segmentation from CT/MRI scans in recent years, a comprehensive evaluation of the models' capabilities is hampered by the lack of a large-scale benchmark from diverse clinical scenarios. Constraint by the high cost of collecting and labeling 3D medical data, most of the deep learning models to date are driven by datasets with a limited number of organs of interest or samples, which still limits the power of modern deep models and makes it difficult to provide a fully comprehensive and fair estimate of various methods. To mitigate the limitations, we present AMOS, a large-scale, diverse, clinical dataset for abdominal organ segmentation. AMOS provides 500 CT and 100 MRI scans collected from multi-center, multi-vendor, multi-modality, multi-phase, multi-disease patients, each with voxel-level annotations of 15 abdominal organs, providing challenging examples and test-bed for studying robust segmentation algorithms under diverse targets and scenarios. We further benchmark several state-of-the-art medical segmentation models to evaluate the status of the existing methods on this new challenging dataset. We have made our datasets, benchmark servers, and baselines publicly available, and hope to inspire future research. Information can be found at https://amos22.grand-challenge.org.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge