Shoujin Huang
Predicting Diabetic Macular Edema Treatment Responses Using OCT: Dataset and Methods of APTOS Competition
May 09, 2025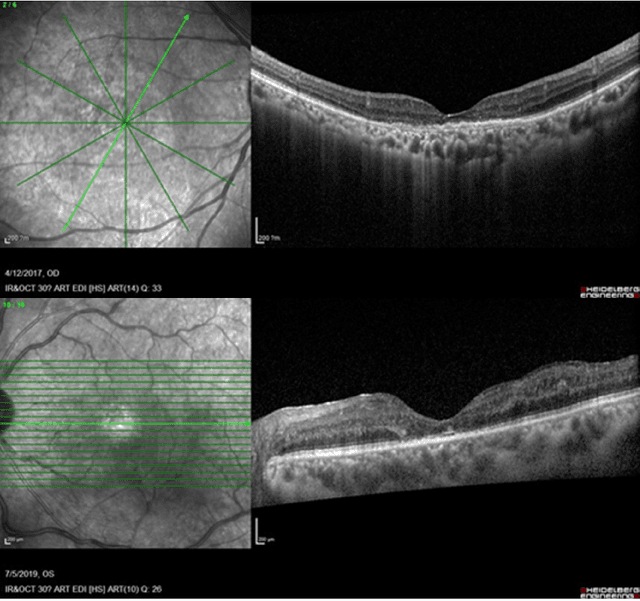
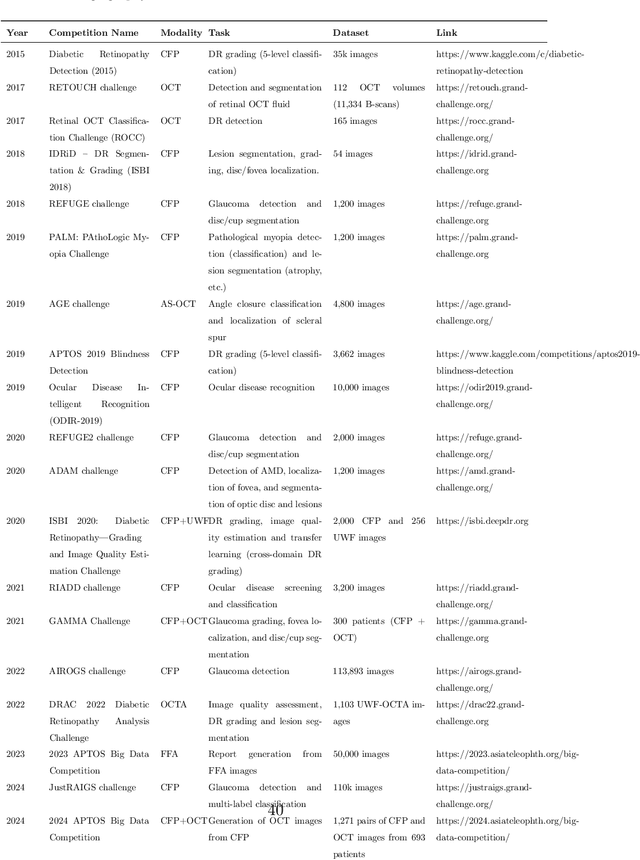
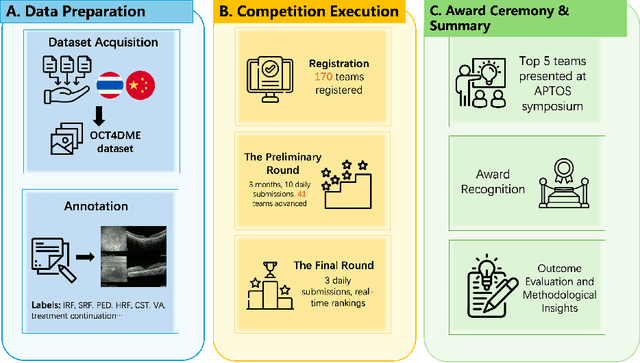

Abstract:Diabetic macular edema (DME) significantly contributes to visual impairment in diabetic patients. Treatment responses to intravitreal therapies vary, highlighting the need for patient stratification to predict therapeutic benefits and enable personalized strategies. To our knowledge, this study is the first to explore pre-treatment stratification for predicting DME treatment responses. To advance this research, we organized the 2nd Asia-Pacific Tele-Ophthalmology Society (APTOS) Big Data Competition in 2021. The competition focused on improving predictive accuracy for anti-VEGF therapy responses using ophthalmic OCT images. We provided a dataset containing tens of thousands of OCT images from 2,000 patients with labels across four sub-tasks. This paper details the competition's structure, dataset, leading methods, and evaluation metrics. The competition attracted strong scientific community participation, with 170 teams initially registering and 41 reaching the final round. The top-performing team achieved an AUC of 80.06%, highlighting the potential of AI in personalized DME treatment and clinical decision-making.
Robust Simultaneous Multislice MRI Reconstruction Using Deep Generative Priors
Jul 31, 2024



Abstract:Simultaneous multislice (SMS) imaging is a powerful technique for accelerating magnetic resonance imaging (MRI) acquisitions. However, SMS reconstruction remains challenging due to the complex signal interactions between and within the excited slices. This study presents a robust SMS MRI reconstruction method using deep generative priors. Starting from Gaussian noise, we leverage denoising diffusion probabilistic models (DDPM) to gradually recover the individual slices through reverse diffusion iterations while imposing data consistency from the measured k-space under readout concatenation framework. The posterior sampling procedure is designed such that the DDPM training can be performed on single-slice images without special adjustments for SMS tasks. Additionally, our method integrates a low-frequency enhancement (LFE) module to address a practical issue that SMS-accelerated fast spin echo (FSE) and echo-planar imaging (EPI) sequences cannot easily embed autocalibration signals. Extensive experiments demonstrate that our approach consistently outperforms existing methods and generalizes well to unseen datasets. The code is available at https://github.com/Solor-pikachu/ROGER after the review process.
Autoregressive Image Diffusion: Generation of Image Sequence and Application in MRI
May 24, 2024



Abstract:Magnetic resonance imaging (MRI) is a widely used non-invasive imaging modality. However, a persistent challenge lies in balancing image quality with imaging speed. This trade-off is primarily constrained by k-space measurements, which traverse specific trajectories in the spatial Fourier domain (k-space). These measurements are often undersampled to shorten acquisition times, resulting in image artifacts and compromised quality. Generative models learn image distributions and can be used to reconstruct high-quality images from undersampled k-space data. In this work, we present the autoregressive image diffusion (AID) model for image sequences and use it to sample the posterior for accelerated MRI reconstruction. The algorithm incorporates both undersampled k-space and pre-existing information. Models trained with fastMRI dataset are evaluated comprehensively. The results show that the AID model can robustly generate sequentially coherent image sequences. In 3D and dynamic MRI, the AID can outperform the standard diffusion model and reduce hallucinations, due to the learned inter-image dependencies.
Autoregressive Image Diffusion: Generating Image Sequence and Application in MRI
May 23, 2024



Abstract:Magnetic resonance imaging (MRI) is a widely used non-invasive imaging modality. However, a persistent challenge lies in balancing image quality with imaging speed. This trade-off is primarily constrained by k-space measurements, which traverse specific trajectories in the spatial Fourier domain (k-space). These measurements are often undersampled to shorten acquisition times, resulting in image artifacts and compromised quality. Generative models learn image distributions and can be used to reconstruct high-quality images from undersampled k-space data. In this work, we present the autoregressive image diffusion (AID) model for image sequences and use it to sample the posterior for accelerated MRI reconstruction. The algorithm incorporates both undersampled k-space and pre-existing information. Models trained with fastMRI dataset are evaluated comprehensively. The results show that the AID model can robustly generate sequentially coherent image sequences. In 3D and dynamic MRI, the AID can outperform the standard diffusion model and reduce hallucinations, due to the learned inter-image dependencies.
Noise Level Adaptive Diffusion Model for Robust Reconstruction of Accelerated MRI
Mar 08, 2024Abstract:In general, diffusion model-based MRI reconstruction methods incrementally remove artificially added noise while imposing data consistency to reconstruct the underlying images. However, real-world MRI acquisitions already contain inherent noise due to thermal fluctuations. This phenomenon is particularly notable when using ultra-fast, high-resolution imaging sequences for advanced research, or using low-field systems favored by low- and middle-income countries. These common scenarios can lead to sub-optimal performance or complete failure of existing diffusion model-based reconstruction techniques. Specifically, as the artificially added noise is gradually removed, the inherent MRI noise becomes increasingly pronounced, making the actual noise level inconsistent with the predefined denoising schedule and consequently inaccurate image reconstruction. To tackle this problem, we propose a posterior sampling strategy with a novel NoIse Level Adaptive Data Consistency (Nila-DC) operation. Extensive experiments are conducted on two public datasets and an in-house clinical dataset with field strength ranging from 0.3T to 3T, showing that our method surpasses the state-of-the-art MRI reconstruction methods, and is highly robust against various noise levels. The code will be released after review.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023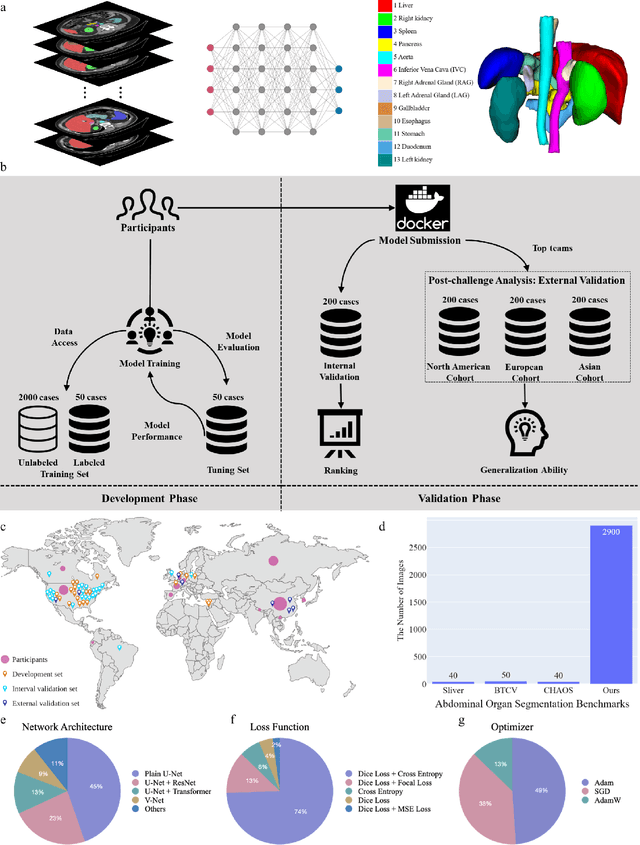
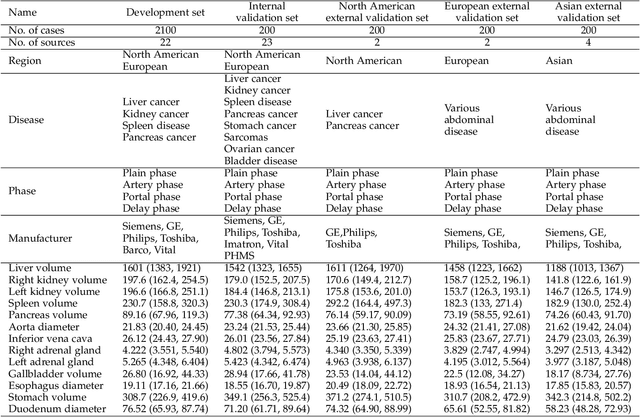
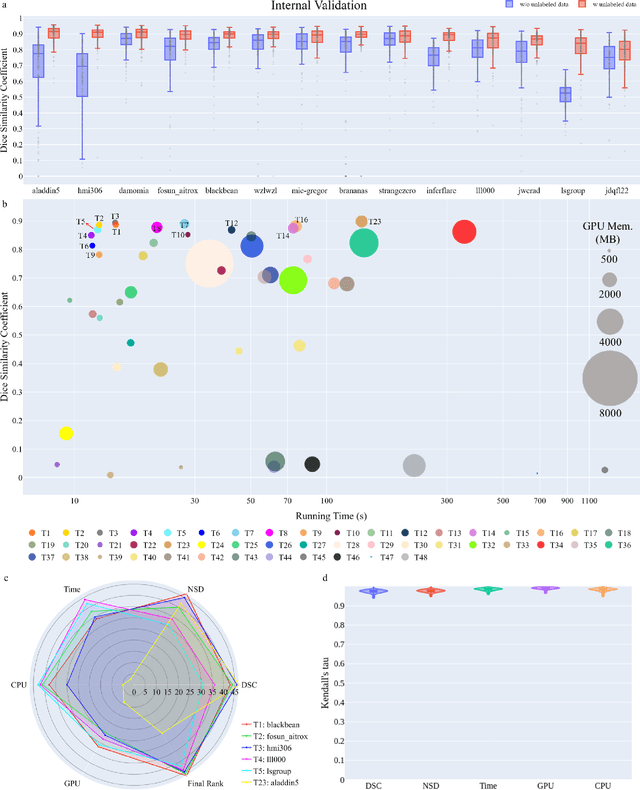
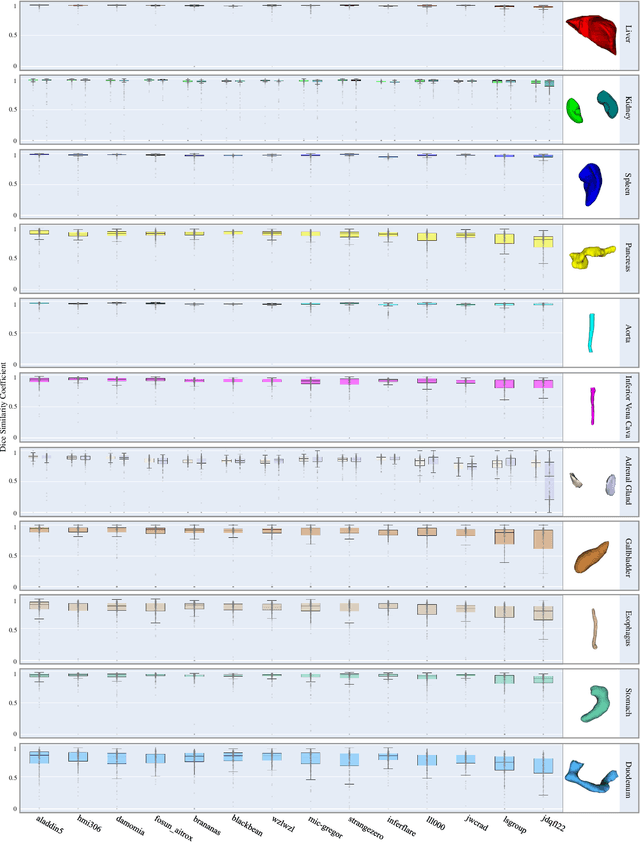
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge