Lina Chen
STEP3-VL-10B Technical Report
Jan 15, 2026Abstract:We present STEP3-VL-10B, a lightweight open-source foundation model designed to redefine the trade-off between compact efficiency and frontier-level multimodal intelligence. STEP3-VL-10B is realized through two strategic shifts: first, a unified, fully unfrozen pre-training strategy on 1.2T multimodal tokens that integrates a language-aligned Perception Encoder with a Qwen3-8B decoder to establish intrinsic vision-language synergy; and second, a scaled post-training pipeline featuring over 1k iterations of reinforcement learning. Crucially, we implement Parallel Coordinated Reasoning (PaCoRe) to scale test-time compute, allocating resources to scalable perceptual reasoning that explores and synthesizes diverse visual hypotheses. Consequently, despite its compact 10B footprint, STEP3-VL-10B rivals or surpasses models 10$\times$-20$\times$ larger (e.g., GLM-4.6V-106B, Qwen3-VL-235B) and top-tier proprietary flagships like Gemini 2.5 Pro and Seed-1.5-VL. Delivering best-in-class performance, it records 92.2% on MMBench and 80.11% on MMMU, while excelling in complex reasoning with 94.43% on AIME2025 and 75.95% on MathVision. We release the full model suite to provide the community with a powerful, efficient, and reproducible baseline.
Progressive Mastery: Customized Curriculum Learning with Guided Prompting for Mathematical Reasoning
Jun 04, 2025Abstract:Large Language Models (LLMs) have achieved remarkable performance across various reasoning tasks, yet post-training is constrained by inefficient sample utilization and inflexible difficulty samples processing. To address these limitations, we propose Customized Curriculum Learning (CCL), a novel framework with two key innovations. First, we introduce model-adaptive difficulty definition that customizes curriculum datasets based on each model's individual capabilities rather than using predefined difficulty metrics. Second, we develop "Guided Prompting," which dynamically reduces sample difficulty through strategic hints, enabling effective utilization of challenging samples that would otherwise degrade performance. Comprehensive experiments on supervised fine-tuning and reinforcement learning demonstrate that CCL significantly outperforms uniform training approaches across five mathematical reasoning benchmarks, confirming its effectiveness across both paradigms in enhancing sample utilization and model performance.
Efficient Self-Supervised Barlow Twins from Limited Tissue Slide Cohorts for Colonic Pathology Diagnostics
Nov 08, 2024



Abstract:Colorectal cancer (CRC) is one of the few cancers that have an established dysplasia-carcinoma sequence that benefits from screening. Everyone over 50 years of age in Canada is eligible for CRC screening. About 20\% of those people will undergo a biopsy for a pre-neoplastic polyp and, in many cases, multiple polyps. As such, these polyp biopsies make up the bulk of a pathologist's workload. Developing an efficient computational model to help screen these polyp biopsies can improve the pathologist's workflow and help guide their attention to critical areas on the slide. DL models face significant challenges in computational pathology (CPath) because of the gigapixel image size of whole-slide images and the scarcity of detailed annotated datasets. It is, therefore, crucial to leverage self-supervised learning (SSL) methods to alleviate the burden and cost of data annotation. However, current research lacks methods to apply SSL frameworks to analyze pathology data effectively. This paper aims to propose an optimized Barlow Twins framework for colorectal polyps screening. We adapt its hyperparameters, augmentation strategy and encoder to the specificity of the pathology data to enhance performance. Additionally, we investigate the best Field of View (FoV) for colorectal polyps screening and propose a new benchmark dataset for CRC screening, made of four types of colorectal polyps and normal tissue, by performing downstream tasking on MHIST and NCT-CRC-7K datasets. Furthermore, we show that the SSL representations are more meaningful and qualitative than the supervised ones and that Barlow Twins benefits from the Swin Transformer when applied to pathology data. Codes are avaialble from https://github.com/AtlasAnalyticsLab/PathBT.
Can Large Language Models Understand Real-World Complex Instructions?
Sep 17, 2023


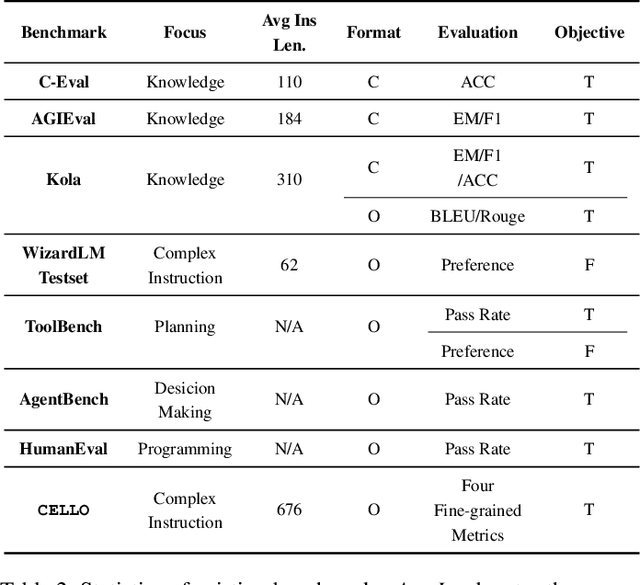
Abstract:Large language models (LLMs) can understand human instructions, showing their potential for pragmatic applications beyond traditional NLP tasks. However, they still struggle with complex instructions, which can be either complex task descriptions that require multiple tasks and constraints, or complex input that contains long context, noise, heterogeneous information and multi-turn format. Due to these features, LLMs often ignore semantic constraints from task descriptions, generate incorrect formats, violate length or sample count constraints, and be unfaithful to the input text. Existing benchmarks are insufficient to assess LLMs' ability to understand complex instructions, as they are close-ended and simple. To bridge this gap, we propose CELLO, a benchmark for evaluating LLMs' ability to follow complex instructions systematically. We design eight features for complex instructions and construct a comprehensive evaluation dataset from real-world scenarios. We also establish four criteria and develop corresponding metrics, as current ones are inadequate, biased or too strict and coarse-grained. We compare the performance of representative Chinese-oriented and English-oriented models in following complex instructions through extensive experiments. Resources of CELLO are publicly available at https://github.com/Abbey4799/CELLO.
HistoKT: Cross Knowledge Transfer in Computational Pathology
Jan 27, 2022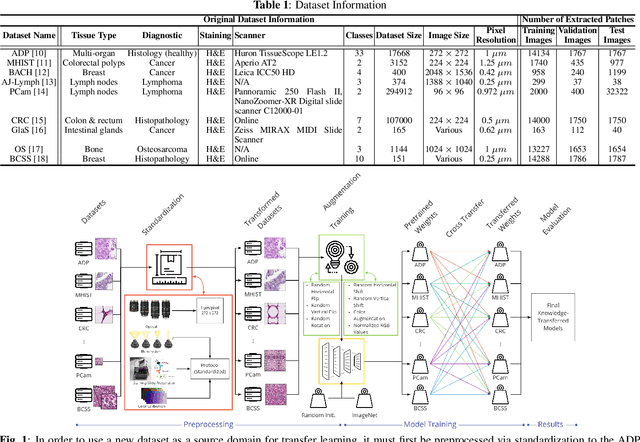



Abstract:The lack of well-annotated datasets in computational pathology (CPath) obstructs the application of deep learning techniques for classifying medical images. %Since pathologist time is expensive, dataset curation is intrinsically difficult. Many CPath workflows involve transferring learned knowledge between various image domains through transfer learning. Currently, most transfer learning research follows a model-centric approach, tuning network parameters to improve transfer results over few datasets. In this paper, we take a data-centric approach to the transfer learning problem and examine the existence of generalizable knowledge between histopathological datasets. First, we create a standardization workflow for aggregating existing histopathological data. We then measure inter-domain knowledge by training ResNet18 models across multiple histopathological datasets, and cross-transferring between them to determine the quantity and quality of innate shared knowledge. Additionally, we use weight distillation to share knowledge between models without additional training. We find that hard to learn, multi-class datasets benefit most from pretraining, and a two stage learning framework incorporating a large source domain such as ImageNet allows for better utilization of smaller datasets. Furthermore, we find that weight distillation enables models trained on purely histopathological features to outperform models using external natural image data.
Probeable DARTS with Application to Computational Pathology
Aug 16, 2021



Abstract:AI technology has made remarkable achievements in computational pathology (CPath), especially with the help of deep neural networks. However, the network performance is highly related to architecture design, which commonly requires human experts with domain knowledge. In this paper, we combat this challenge with the recent advance in neural architecture search (NAS) to find an optimal network for CPath applications. In particular, we use differentiable architecture search (DARTS) for its efficiency. We first adopt a probing metric to show that the original DARTS lacks proper hyperparameter tuning on the CIFAR dataset, and how the generalization issue can be addressed using an adaptive optimization strategy. We then apply our searching framework on CPath applications by searching for the optimum network architecture on a histological tissue type dataset (ADP). Results show that the searched network outperforms state-of-the-art networks in terms of prediction accuracy and computation complexity. We further conduct extensive experiments to demonstrate the transferability of the searched network to new CPath applications, the robustness against downscaled inputs, as well as the reliability of predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge