Ryan Zhang
Dynamically Learned Test-Time Model Routing in Language Model Zoos with Service Level Guarantees
May 26, 2025Abstract:Open-weight LLM zoos provide access to numerous high-quality models, but selecting the appropriate model for specific tasks remains challenging and requires technical expertise. Most users simply want factually correct, safe, and satisfying responses without concerning themselves with model technicalities, while inference service providers prioritize minimizing operating costs. These competing interests are typically mediated through service level agreements (SLAs) that guarantee minimum service quality. We introduce MESS+, a stochastic optimization algorithm for cost-optimal LLM request routing while providing rigorous SLA compliance guarantees. MESS+ learns request satisfaction probabilities of LLMs in real-time as users interact with the system, based on which model selection decisions are made by solving a per-request optimization problem. Our algorithm includes a novel combination of virtual queues and request satisfaction prediction, along with a theoretical analysis of cost optimality and constraint satisfaction. Across a wide range of state-of-the-art LLM benchmarks, MESS+ achieves an average of 2x cost savings compared to existing LLM routing techniques.
MESS+: Energy-Optimal Inferencing in Language Model Zoos with Service Level Guarantees
Oct 31, 2024



Abstract:Open-weight large language model (LLM) zoos allow users to quickly integrate state-of-the-art models into systems. Despite increasing availability, selecting the most appropriate model for a given task still largely relies on public benchmark leaderboards and educated guesses. This can be unsatisfactory for both inference service providers and end users, where the providers usually prioritize cost efficiency, while the end users usually prioritize model output quality for their inference requests. In commercial settings, these two priorities are often brought together in Service Level Agreements (SLA). We present MESS+, an online stochastic optimization algorithm for energy-optimal model selection from a model zoo, which works on a per-inference-request basis. For a given SLA that requires high accuracy, we are up to 2.5x more energy efficient with MESS+ than with randomly selecting an LLM from the zoo while maintaining SLA quality constraints.
LiteGPT: Large Vision-Language Model for Joint Chest X-ray Localization and Classification Task
Jul 16, 2024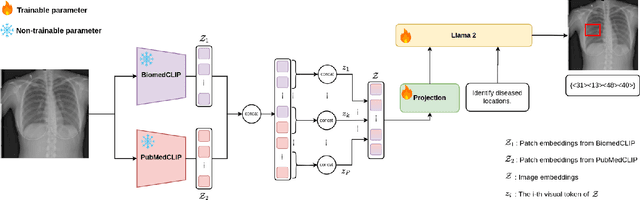
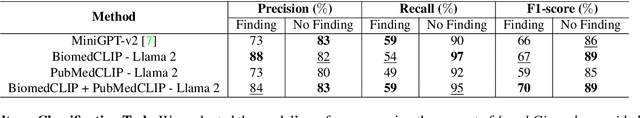
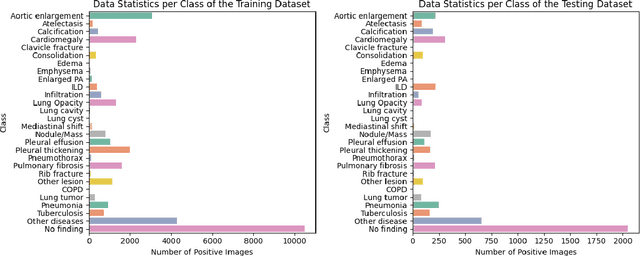
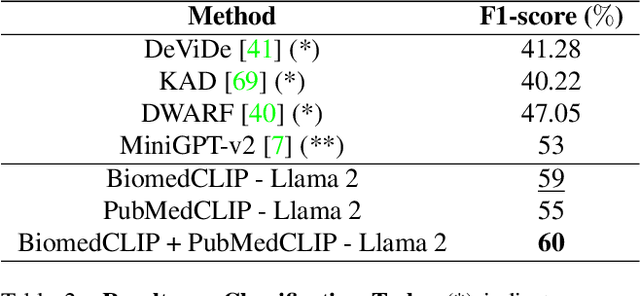
Abstract:Vision-language models have been extensively explored across a wide range of tasks, achieving satisfactory performance; however, their application in medical imaging remains underexplored. In this work, we propose a unified framework - LiteGPT - for the medical imaging. We leverage multiple pre-trained visual encoders to enrich information and enhance the performance of vision-language models. To the best of our knowledge, this is the first study to utilize vision-language models for the novel task of joint localization and classification in medical images. Besides, we are pioneers in providing baselines for disease localization in chest X-rays. Finally, we set new state-of-the-art performance in the image classification task on the well-benchmarked VinDr-CXR dataset. All code and models are publicly available online: https://github.com/leduckhai/LiteGPT
Computational Pathology: A Survey Review and The Way Forward
Apr 11, 2023



Abstract:Computational Pathology (CoPath) is an interdisciplinary science that augments developments of computational approaches to analyze and model medical histopathology images. The main objective for CoPath is to develop infrastructure and workflows of digital diagnostics as an assistive CAD system for clinical pathology facilitating transformational changes in the diagnosis and treatment of cancer diseases. With evergrowing developments in deep learning and computer vision algorithms, and the ease of the data flow from digital pathology, currently CoPath is witnessing a paradigm shift. Despite the sheer volume of engineering and scientific works being introduced for cancer image analysis, there is still a considerable gap of adopting and integrating these algorithms in clinical practice. This raises a significant question regarding the direction and trends that are undertaken in CoPath. In this article we provide a comprehensive review of more than 700 papers to address the challenges faced in problem design all-the-way to the application and implementation viewpoints. We have catalogued each paper into a model-card by examining the key works and challenges faced to layout the current landscape in CoPath. We hope this helps the community to locate relevant works and facilitate understanding of the field's future directions. In a nutshell, we oversee the CoPath developments in cycle of stages which are required to be cohesively linked together to address the challenges associated with such multidisciplinary science. We overview this cycle from different perspectives of data-centric, model-centric, and application-centric problems. We finally sketch remaining challenges and provide directions for future technical developments and clinical integration of CoPath.
HistoKT: Cross Knowledge Transfer in Computational Pathology
Jan 27, 2022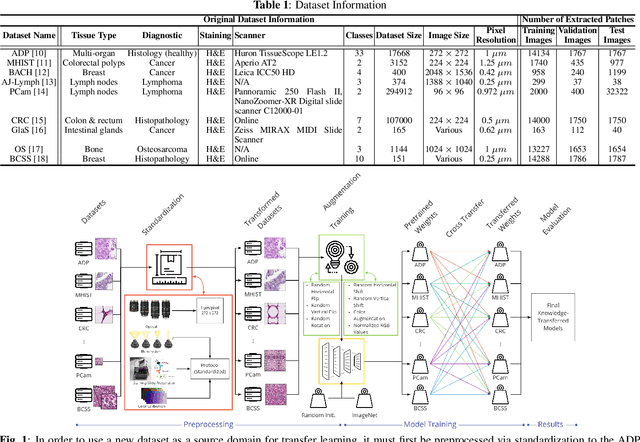



Abstract:The lack of well-annotated datasets in computational pathology (CPath) obstructs the application of deep learning techniques for classifying medical images. %Since pathologist time is expensive, dataset curation is intrinsically difficult. Many CPath workflows involve transferring learned knowledge between various image domains through transfer learning. Currently, most transfer learning research follows a model-centric approach, tuning network parameters to improve transfer results over few datasets. In this paper, we take a data-centric approach to the transfer learning problem and examine the existence of generalizable knowledge between histopathological datasets. First, we create a standardization workflow for aggregating existing histopathological data. We then measure inter-domain knowledge by training ResNet18 models across multiple histopathological datasets, and cross-transferring between them to determine the quantity and quality of innate shared knowledge. Additionally, we use weight distillation to share knowledge between models without additional training. We find that hard to learn, multi-class datasets benefit most from pretraining, and a two stage learning framework incorporating a large source domain such as ImageNet allows for better utilization of smaller datasets. Furthermore, we find that weight distillation enables models trained on purely histopathological features to outperform models using external natural image data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge