Stephen Yang
DETECT: Data-Driven Evaluation of Treatments Enabled by Classification Transformers
Nov 10, 2025Abstract:Chronic pain is a global health challenge affecting millions of individuals, making it essential for physicians to have reliable and objective methods to measure the functional impact of clinical treatments. Traditionally used methods, like the numeric rating scale, while personalized and easy to use, are subjective due to their self-reported nature. Thus, this paper proposes DETECT (Data-Driven Evaluation of Treatments Enabled by Classification Transformers), a data-driven framework that assesses treatment success by comparing patient activities of daily life before and after treatment. We use DETECT on public benchmark datasets and simulated patient data from smartphone sensors. Our results demonstrate that DETECT is objective yet lightweight, making it a significant and novel contribution to clinical decision-making. By using DETECT, independently or together with other self-reported metrics, physicians can improve their understanding of their treatment impacts, ultimately leading to more personalized and responsive patient care.
VibES: Induced Vibration for Persistent Event-Based Sensing
Aug 26, 2025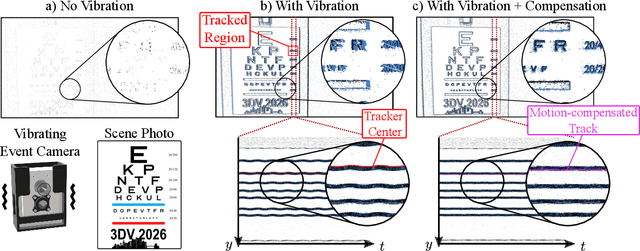
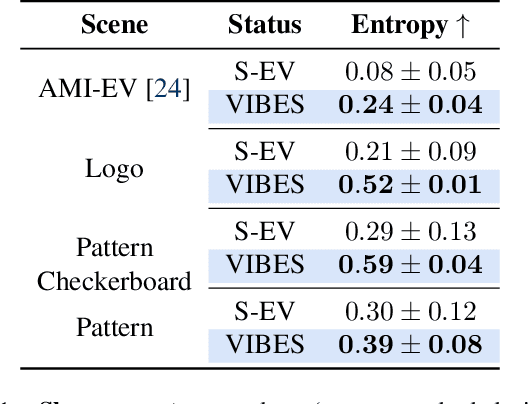
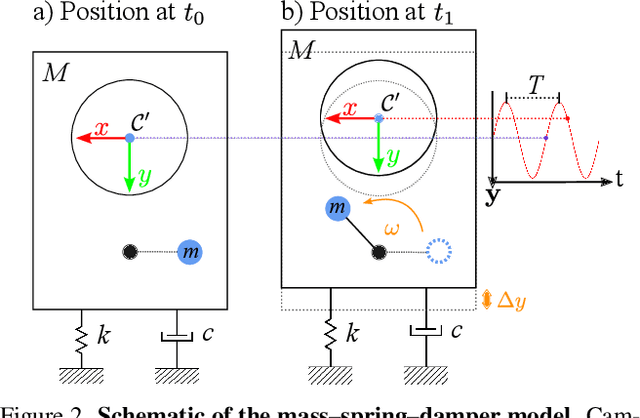
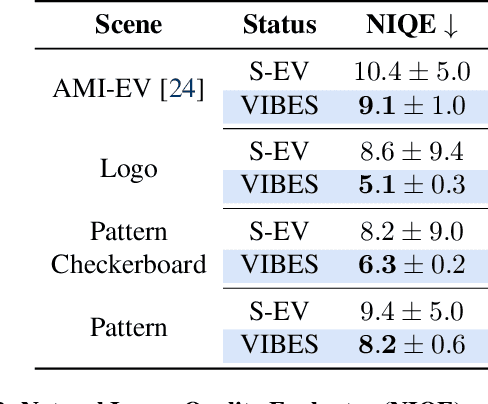
Abstract:Event cameras are a bio-inspired class of sensors that asynchronously measure per-pixel intensity changes. Under fixed illumination conditions in static or low-motion scenes, rigidly mounted event cameras are unable to generate any events, becoming unsuitable for most computer vision tasks. To address this limitation, recent work has investigated motion-induced event stimulation that often requires complex hardware or additional optical components. In contrast, we introduce a lightweight approach to sustain persistent event generation by employing a simple rotating unbalanced mass to induce periodic vibrational motion. This is combined with a motion-compensation pipeline that removes the injected motion and yields clean, motion-corrected events for downstream perception tasks. We demonstrate our approach with a hardware prototype and evaluate it on real-world captured datasets. Our method reliably recovers motion parameters and improves both image reconstruction and edge detection over event-based sensing without motion induction.
The Participatory Turn in AI Design: Theoretical Foundations and the Current State of Practice
Oct 02, 2023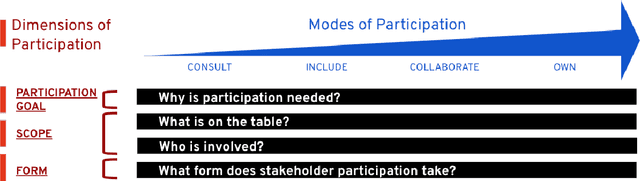
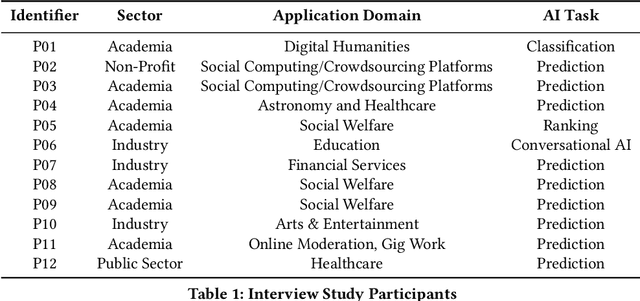
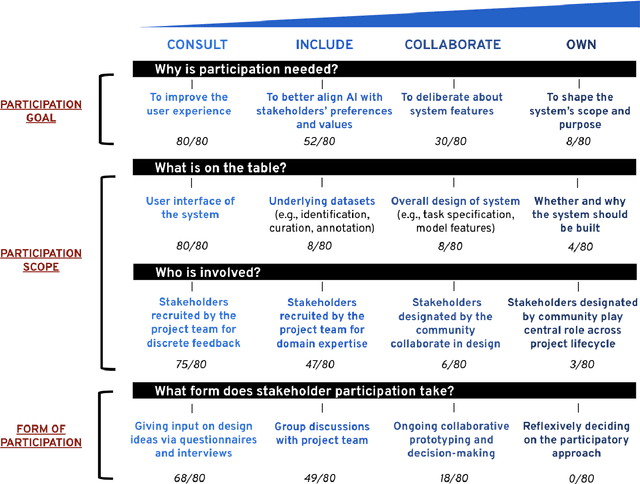
Abstract:Despite the growing consensus that stakeholders affected by AI systems should participate in their design, enormous variation and implicit disagreements exist among current approaches. For researchers and practitioners who are interested in taking a participatory approach to AI design and development, it remains challenging to assess the extent to which any participatory approach grants substantive agency to stakeholders. This article thus aims to ground what we dub the "participatory turn" in AI design by synthesizing existing theoretical literature on participation and through empirical investigation and critique of its current practices. Specifically, we derive a conceptual framework through synthesis of literature across technology design, political theory, and the social sciences that researchers and practitioners can leverage to evaluate approaches to participation in AI design. Additionally, we articulate empirical findings concerning the current state of participatory practice in AI design based on an analysis of recently published research and semi-structured interviews with 12 AI researchers and practitioners. We use these empirical findings to understand the current state of participatory practice and subsequently provide guidance to better align participatory goals and methods in a way that accounts for practical constraints.
Predictive Patentomics: Forecasting Innovation Success and Valuation with ChatGPT
Jun 22, 2023Abstract:Analysis of innovation has been fundamentally limited by conventional approaches to broad, structural variables. This paper pushes the boundaries, taking an LLM approach to patent analysis with the groundbreaking ChatGPT technology. OpenAI's state-of-the-art textual embedding accesses complex information about the quality and impact of each invention to power deep learning predictive models. The nuanced embedding drives a 24% incremental improvement in R-squared predicting patent value and clearly isolates the worst and best applications. These models enable a revision of the contemporary Kogan, Papanikolaou, Seru, and Stoffman (2017) valuation of patents by a median deviation of 1.5 times, accounting for potential institutional predictions. Furthermore, the market fails to incorporate timely information about applications; a long-short portfolio based on predicted acceptance rates achieves significant abnormal returns of 3.3% annually. The models provide an opportunity to revolutionize startup and small-firm corporate policy vis-a-vis patenting.
Computational Pathology: A Survey Review and The Way Forward
Apr 11, 2023



Abstract:Computational Pathology (CoPath) is an interdisciplinary science that augments developments of computational approaches to analyze and model medical histopathology images. The main objective for CoPath is to develop infrastructure and workflows of digital diagnostics as an assistive CAD system for clinical pathology facilitating transformational changes in the diagnosis and treatment of cancer diseases. With evergrowing developments in deep learning and computer vision algorithms, and the ease of the data flow from digital pathology, currently CoPath is witnessing a paradigm shift. Despite the sheer volume of engineering and scientific works being introduced for cancer image analysis, there is still a considerable gap of adopting and integrating these algorithms in clinical practice. This raises a significant question regarding the direction and trends that are undertaken in CoPath. In this article we provide a comprehensive review of more than 700 papers to address the challenges faced in problem design all-the-way to the application and implementation viewpoints. We have catalogued each paper into a model-card by examining the key works and challenges faced to layout the current landscape in CoPath. We hope this helps the community to locate relevant works and facilitate understanding of the field's future directions. In a nutshell, we oversee the CoPath developments in cycle of stages which are required to be cohesively linked together to address the challenges associated with such multidisciplinary science. We overview this cycle from different perspectives of data-centric, model-centric, and application-centric problems. We finally sketch remaining challenges and provide directions for future technical developments and clinical integration of CoPath.
HistoKT: Cross Knowledge Transfer in Computational Pathology
Jan 27, 2022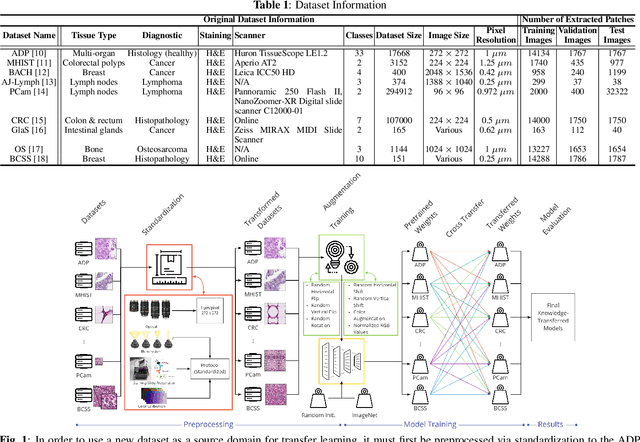



Abstract:The lack of well-annotated datasets in computational pathology (CPath) obstructs the application of deep learning techniques for classifying medical images. %Since pathologist time is expensive, dataset curation is intrinsically difficult. Many CPath workflows involve transferring learned knowledge between various image domains through transfer learning. Currently, most transfer learning research follows a model-centric approach, tuning network parameters to improve transfer results over few datasets. In this paper, we take a data-centric approach to the transfer learning problem and examine the existence of generalizable knowledge between histopathological datasets. First, we create a standardization workflow for aggregating existing histopathological data. We then measure inter-domain knowledge by training ResNet18 models across multiple histopathological datasets, and cross-transferring between them to determine the quantity and quality of innate shared knowledge. Additionally, we use weight distillation to share knowledge between models without additional training. We find that hard to learn, multi-class datasets benefit most from pretraining, and a two stage learning framework incorporating a large source domain such as ImageNet allows for better utilization of smaller datasets. Furthermore, we find that weight distillation enables models trained on purely histopathological features to outperform models using external natural image data.
Stakeholder Participation in AI: Beyond "Add Diverse Stakeholders and Stir"
Nov 01, 2021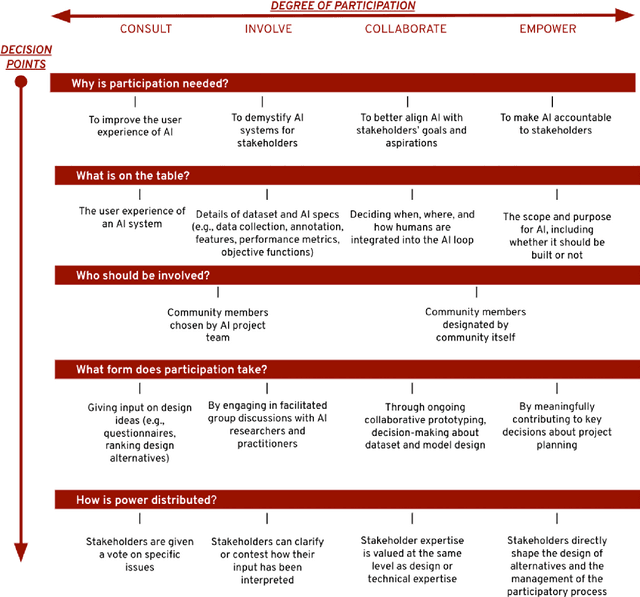
Abstract:There is a growing consensus in HCI and AI research that the design of AI systems needs to engage and empower stakeholders who will be affected by AI. However, the manner in which stakeholders should participate in AI design is unclear. This workshop paper aims to ground what we dub a 'participatory turn' in AI design by synthesizing existing literature on participation and through empirical analysis of its current practices via a survey of recent published research and a dozen semi-structured interviews with AI researchers and practitioners. Based on our literature synthesis and empirical research, this paper presents a conceptual framework for analyzing participatory approaches to AI design and articulates a set of empirical findings that in ensemble detail out the contemporary landscape of participatory practice in AI design. These findings can help bootstrap a more principled discussion on how PD of AI should move forward across AI, HCI, and other research communities.
Clinical Evidence Engine: Proof-of-Concept For A Clinical-Domain-Agnostic Decision Support Infrastructure
Oct 31, 2021



Abstract:Abstruse learning algorithms and complex datasets increasingly characterize modern clinical decision support systems (CDSS). As a result, clinicians cannot easily or rapidly scrutinize the CDSS recommendation when facing a difficult diagnosis or treatment decision in practice. Over-trust or under-trust are frequent. Prior research has explored supporting such assessments by explaining DST data inputs and algorithmic mechanisms. This paper explores a different approach: Providing precisely relevant, scientific evidence from biomedical literature. We present a proof-of-concept system, Clinical Evidence Engine, to demonstrate the technical and design feasibility of this approach across three domains (cardiovascular diseases, autism, cancer). Leveraging Clinical BioBERT, the system can effectively identify clinical trial reports based on lengthy clinical questions (e.g., "risks of catheter infection among adult patients in intensive care unit who require arterial catheters, if treated with povidone iodine-alcohol"). This capability enables the system to identify clinical trials relevant to diagnostic/treatment hypotheses -- a clinician's or a CDSS's. Further, Clinical Evidence Engine can identify key parts of a clinical trial abstract, including patient population (e.g., adult patients in intensive care unit who require arterial catheters), intervention (povidone iodine-alcohol), and outcome (risks of catheter infection). This capability opens up the possibility of enabling clinicians to 1) rapidly determine the match between a clinical trial and a clinical question, and 2) understand the result and contexts of the trial without extensive reading. We demonstrate this potential by illustrating two example use scenarios of the system. We discuss the idea of designing DST explanations not as specific to a DST or an algorithm, but as a domain-agnostic decision support infrastructure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge