Vasudev Sharma
Investigating Zero-Shot Diagnostic Pathology in Vision-Language Models with Efficient Prompt Design
Apr 30, 2025



Abstract:Vision-language models (VLMs) have gained significant attention in computational pathology due to their multimodal learning capabilities that enhance big-data analytics of giga-pixel whole slide image (WSI). However, their sensitivity to large-scale clinical data, task formulations, and prompt design remains an open question, particularly in terms of diagnostic accuracy. In this paper, we present a systematic investigation and analysis of three state of the art VLMs for histopathology, namely Quilt-Net, Quilt-LLAVA, and CONCH, on an in-house digestive pathology dataset comprising 3,507 WSIs, each in giga-pixel form, across distinct tissue types. Through a structured ablative study on cancer invasiveness and dysplasia status, we develop a comprehensive prompt engineering framework that systematically varies domain specificity, anatomical precision, instructional framing, and output constraints. Our findings demonstrate that prompt engineering significantly impacts model performance, with the CONCH model achieving the highest accuracy when provided with precise anatomical references. Additionally, we identify the critical importance of anatomical context in histopathological image analysis, as performance consistently degraded when reducing anatomical precision. We also show that model complexity alone does not guarantee superior performance, as effective domain alignment and domain-specific training are critical. These results establish foundational guidelines for prompt engineering in computational pathology and highlight the potential of VLMs to enhance diagnostic accuracy when properly instructed with domain-appropriate prompts.
Efficient Self-Supervised Barlow Twins from Limited Tissue Slide Cohorts for Colonic Pathology Diagnostics
Nov 08, 2024



Abstract:Colorectal cancer (CRC) is one of the few cancers that have an established dysplasia-carcinoma sequence that benefits from screening. Everyone over 50 years of age in Canada is eligible for CRC screening. About 20\% of those people will undergo a biopsy for a pre-neoplastic polyp and, in many cases, multiple polyps. As such, these polyp biopsies make up the bulk of a pathologist's workload. Developing an efficient computational model to help screen these polyp biopsies can improve the pathologist's workflow and help guide their attention to critical areas on the slide. DL models face significant challenges in computational pathology (CPath) because of the gigapixel image size of whole-slide images and the scarcity of detailed annotated datasets. It is, therefore, crucial to leverage self-supervised learning (SSL) methods to alleviate the burden and cost of data annotation. However, current research lacks methods to apply SSL frameworks to analyze pathology data effectively. This paper aims to propose an optimized Barlow Twins framework for colorectal polyps screening. We adapt its hyperparameters, augmentation strategy and encoder to the specificity of the pathology data to enhance performance. Additionally, we investigate the best Field of View (FoV) for colorectal polyps screening and propose a new benchmark dataset for CRC screening, made of four types of colorectal polyps and normal tissue, by performing downstream tasking on MHIST and NCT-CRC-7K datasets. Furthermore, we show that the SSL representations are more meaningful and qualitative than the supervised ones and that Barlow Twins benefits from the Swin Transformer when applied to pathology data. Codes are avaialble from https://github.com/AtlasAnalyticsLab/PathBT.
Masked Autoencoders for Microscopy are Scalable Learners of Cellular Biology
Apr 16, 2024



Abstract:Featurizing microscopy images for use in biological research remains a significant challenge, especially for large-scale experiments spanning millions of images. This work explores the scaling properties of weakly supervised classifiers and self-supervised masked autoencoders (MAEs) when training with increasingly larger model backbones and microscopy datasets. Our results show that ViT-based MAEs outperform weakly supervised classifiers on a variety of tasks, achieving as much as a 11.5% relative improvement when recalling known biological relationships curated from public databases. Additionally, we develop a new channel-agnostic MAE architecture (CA-MAE) that allows for inputting images of different numbers and orders of channels at inference time. We demonstrate that CA-MAEs effectively generalize by inferring and evaluating on a microscopy image dataset (JUMP-CP) generated under different experimental conditions with a different channel structure than our pretraining data (RPI-93M). Our findings motivate continued research into scaling self-supervised learning on microscopy data in order to create powerful foundation models of cellular biology that have the potential to catalyze advancements in drug discovery and beyond.
Masked autoencoders are scalable learners of cellular morphology
Sep 27, 2023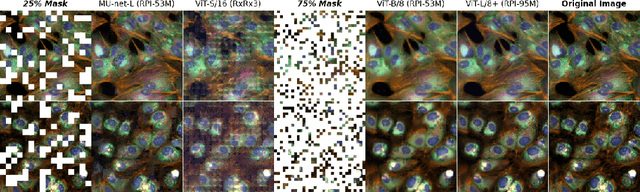
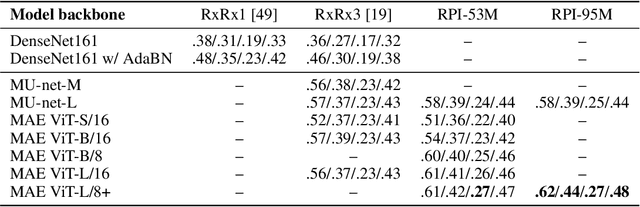
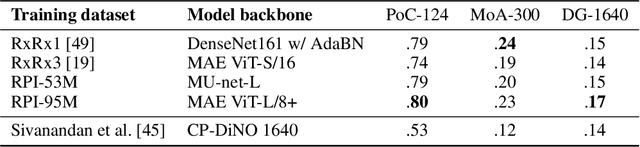
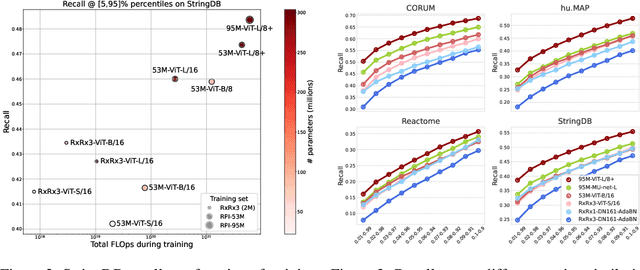
Abstract:Inferring biological relationships from cellular phenotypes in high-content microscopy screens provides significant opportunity and challenge in biological research. Prior results have shown that deep vision models can capture biological signal better than hand-crafted features. This work explores how weakly supervised and self-supervised deep learning approaches scale when training larger models on larger datasets. Our results show that both CNN- and ViT-based masked autoencoders significantly outperform weakly supervised models. At the high-end of our scale, a ViT-L/8 trained on over 3.5-billion unique crops sampled from 95-million microscopy images achieves relative improvements as high as 28% over our best weakly supervised models at inferring known biological relationships curated from public databases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge