Junheng Hao
CUA-Skill: Develop Skills for Computer Using Agent
Jan 28, 2026Abstract:Computer-Using Agents (CUAs) aim to autonomously operate computer systems to complete real-world tasks. However, existing agentic systems remain difficult to scale and lag behind human performance. A key limitation is the absence of reusable and structured skill abstractions that capture how humans interact with graphical user interfaces and how to leverage these skills. We introduce CUA-Skill, a computer-using agentic skill base that encodes human computer-use knowledge as skills coupled with parameterized execution and composition graphs. CUA-Skill is a large-scale library of carefully engineered skills spanning common Windows applications, serving as a practical infrastructure and tool substrate for scalable, reliable agent development. Built upon this skill base, we construct CUA-Skill Agent, an end-to-end computer-using agent that supports dynamic skill retrieval, argument instantiation, and memory-aware failure recovery. Our results demonstrate that CUA-Skill substantially improves execution success rates and robustness on challenging end-to-end agent benchmarks, establishing a strong foundation for future computer-using agent development. On WindowsAgentArena, CUA-Skill Agent achieves state-of-the-art 57.5% (best of three) successful rate while being significantly more efficient than prior and concurrent approaches. The project page is available at https://microsoft.github.io/cua_skill/.
Phi-4-Mini Technical Report: Compact yet Powerful Multimodal Language Models via Mixture-of-LoRAs
Mar 03, 2025



Abstract:We introduce Phi-4-Mini and Phi-4-Multimodal, compact yet highly capable language and multimodal models. Phi-4-Mini is a 3.8-billion-parameter language model trained on high-quality web and synthetic data, significantly outperforming recent open-source models of similar size and matching the performance of models twice its size on math and coding tasks requiring complex reasoning. This achievement is driven by a carefully curated synthetic data recipe emphasizing high-quality math and coding datasets. Compared to its predecessor, Phi-3.5-Mini, Phi-4-Mini features an expanded vocabulary size of 200K tokens to better support multilingual applications, as well as group query attention for more efficient long-sequence generation. Phi-4-Multimodal is a multimodal model that integrates text, vision, and speech/audio input modalities into a single model. Its novel modality extension approach leverages LoRA adapters and modality-specific routers to allow multiple inference modes combining various modalities without interference. For example, it now ranks first in the OpenASR leaderboard to date, although the LoRA component of the speech/audio modality has just 460 million parameters. Phi-4-Multimodal supports scenarios involving (vision + language), (vision + speech), and (speech/audio) inputs, outperforming larger vision-language and speech-language models on a wide range of tasks. Additionally, we experiment to further train Phi-4-Mini to enhance its reasoning capabilities. Despite its compact 3.8-billion-parameter size, this experimental version achieves reasoning performance on par with or surpassing significantly larger models, including DeepSeek-R1-Distill-Qwen-7B and DeepSeek-R1-Distill-Llama-8B.
Phi-3 Technical Report: A Highly Capable Language Model Locally on Your Phone
Apr 23, 2024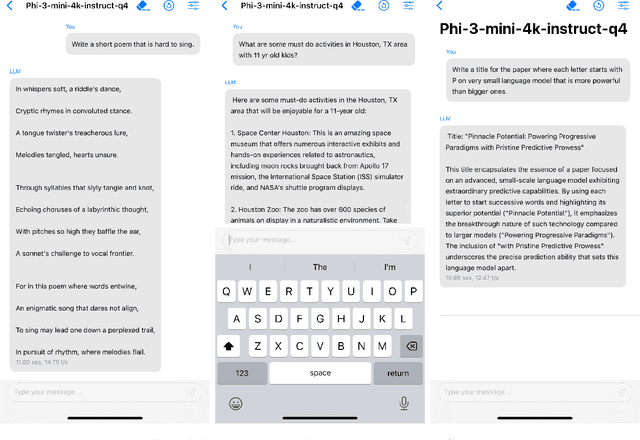
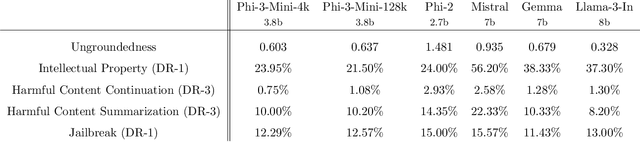
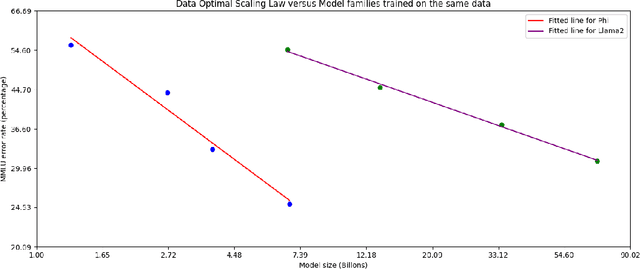
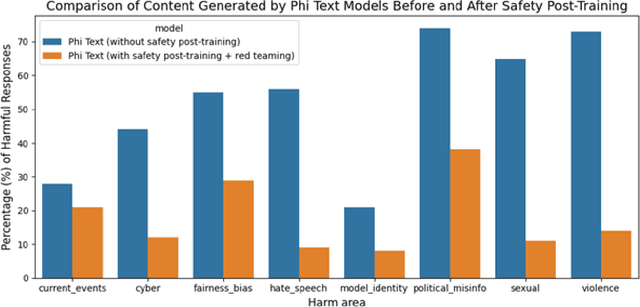
Abstract:We introduce phi-3-mini, a 3.8 billion parameter language model trained on 3.3 trillion tokens, whose overall performance, as measured by both academic benchmarks and internal testing, rivals that of models such as Mixtral 8x7B and GPT-3.5 (e.g., phi-3-mini achieves 69% on MMLU and 8.38 on MT-bench), despite being small enough to be deployed on a phone. The innovation lies entirely in our dataset for training, a scaled-up version of the one used for phi-2, composed of heavily filtered web data and synthetic data. The model is also further aligned for robustness, safety, and chat format. We also provide some initial parameter-scaling results with a 7B and 14B models trained for 4.8T tokens, called phi-3-small and phi-3-medium, both significantly more capable than phi-3-mini (e.g., respectively 75% and 78% on MMLU, and 8.7 and 8.9 on MT-bench).
SciAgent: Tool-augmented Language Models for Scientific Reasoning
Feb 21, 2024



Abstract:Scientific reasoning poses an excessive challenge for even the most advanced Large Language Models (LLMs). To make this task more practical and solvable for LLMs, we introduce a new task setting named tool-augmented scientific reasoning. This setting supplements LLMs with scalable toolsets, and shifts the focus from pursuing an omniscient problem solver to a proficient tool-user. To facilitate the research of such setting, we construct a tool-augmented training corpus named MathFunc which encompasses over 30,000 samples and roughly 6,000 tools. Building on MathFunc, we develop SciAgent to retrieve, understand and, if necessary, use tools for scientific problem solving. Additionally, we craft a benchmark, SciToolBench, spanning five scientific domains to evaluate LLMs' abilities with tool assistance. Extensive experiments on SciToolBench confirm the effectiveness of SciAgent. Notably, SciAgent-Mistral-7B surpasses other LLMs with the same size by more than 13% in absolute accuracy. Furthermore, SciAgent-DeepMath-7B shows much superior performance than ChatGPT.
Language Models can be Logical Solvers
Nov 10, 2023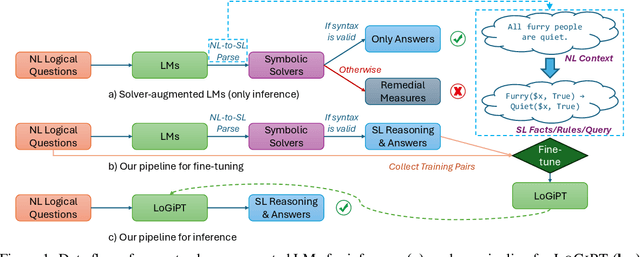
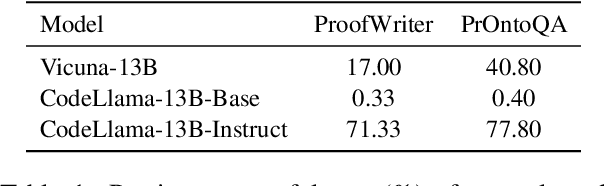
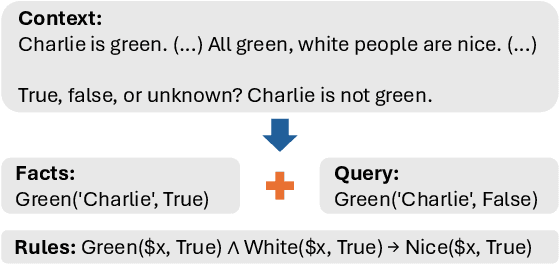
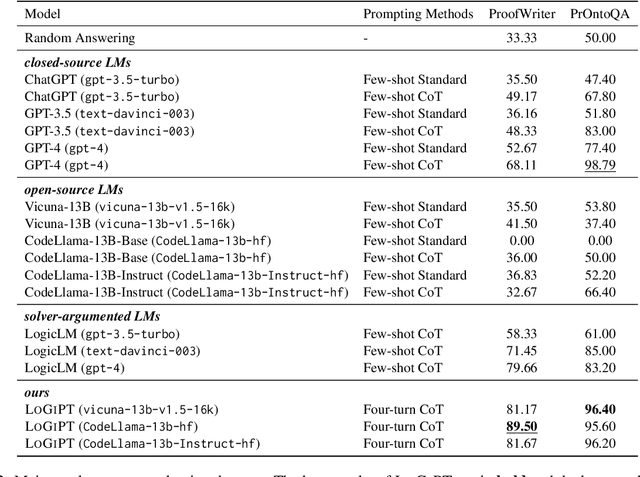
Abstract:Logical reasoning is a fundamental aspect of human intelligence and a key component of tasks like problem-solving and decision-making. Recent advancements have enabled Large Language Models (LLMs) to potentially exhibit reasoning capabilities, but complex logical reasoning remains a challenge. The state-of-the-art, solver-augmented language models, use LLMs to parse natural language logical questions into symbolic representations first and then adopt external logical solvers to take in the symbolic representations and output the answers. Despite their impressive performance, any parsing errors will inevitably result in the failure of the execution of the external logical solver and no answer to the logical questions. In this paper, we introduce LoGiPT, a novel language model that directly emulates the reasoning processes of logical solvers and bypasses the parsing errors by learning to strict adherence to solver syntax and grammar. LoGiPT is fine-tuned on a newly constructed instruction-tuning dataset derived from revealing and refining the invisible reasoning process of deductive solvers. Experimental results on two public deductive reasoning datasets demonstrate that LoGiPT outperforms state-of-the-art solver-augmented LMs and few-shot prompting methods on competitive LLMs like ChatGPT or GPT-4.
A Mobility-Aware Deep Learning Model for Long-Term COVID-19 Pandemic Prediction and Policy Impact Analysis
Dec 05, 2022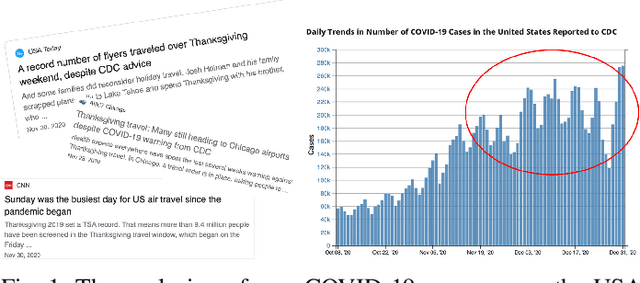
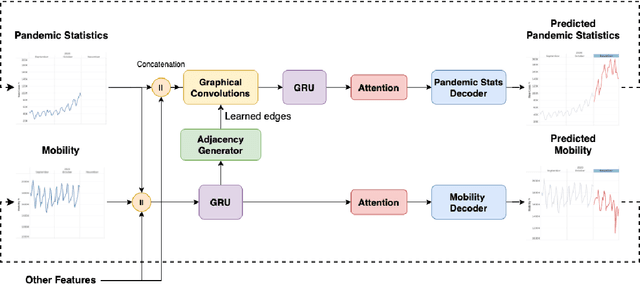
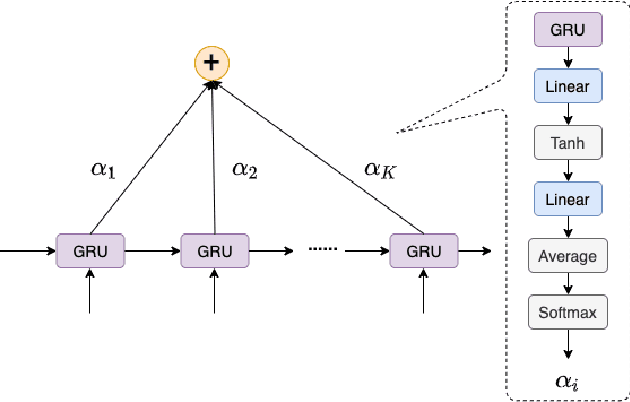
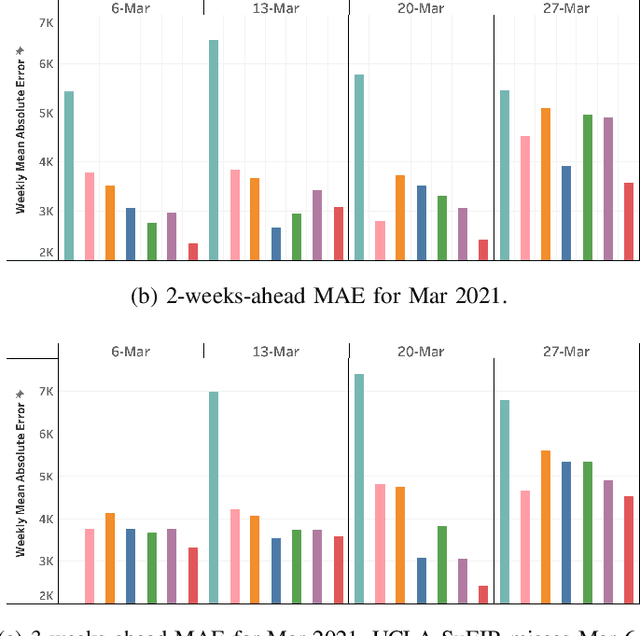
Abstract:Pandemic(epidemic) modeling, aiming at disease spreading analysis, has always been a popular research topic especially following the outbreak of COVID-19 in 2019. Some representative models including SIR-based deep learning prediction models have shown satisfactory performance. However, one major drawback for them is that they fall short in their long-term predictive ability. Although graph convolutional networks (GCN) also perform well, their edge representations do not contain complete information and it can lead to biases. Another drawback is that they usually use input features which they are unable to predict. Hence, those models are unable to predict further future. We propose a model that can propagate predictions further into the future and it has better edge representations. In particular, we model the pandemic as a spatial-temporal graph whose edges represent the transition of infections and are learned by our model. We use a two-stream framework that contains GCN and recursive structures (GRU) with an attention mechanism. Our model enables mobility analysis that provides an effective toolbox for public health researchers and policy makers to predict how different lock-down strategies that actively control mobility can influence the spread of pandemics. Experiments show that our model outperforms others in its long-term predictive power. Moreover, we simulate the effects of certain policies and predict their impacts on infection control.
Metadata-Induced Contrastive Learning for Zero-Shot Multi-Label Text Classification
Feb 11, 2022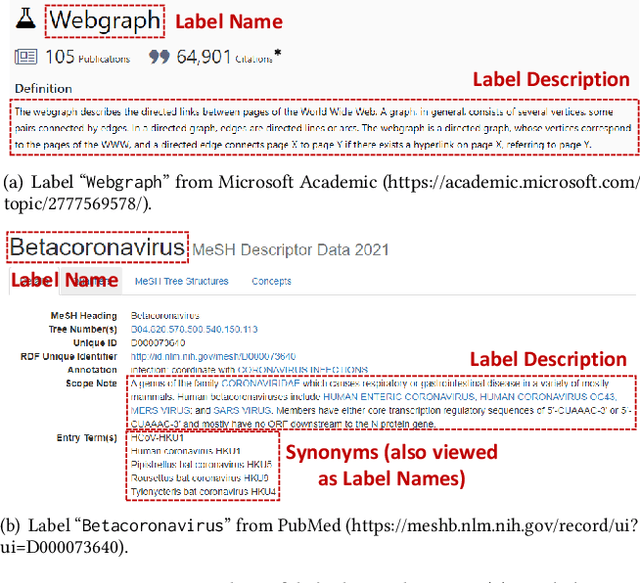
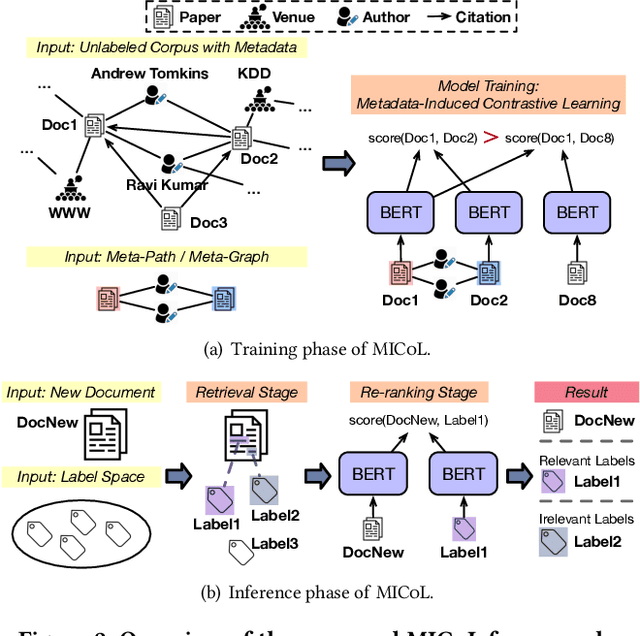
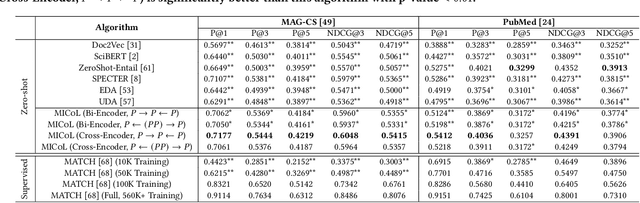
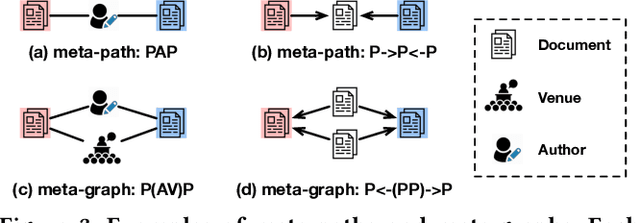
Abstract:Large-scale multi-label text classification (LMTC) aims to associate a document with its relevant labels from a large candidate set. Most existing LMTC approaches rely on massive human-annotated training data, which are often costly to obtain and suffer from a long-tailed label distribution (i.e., many labels occur only a few times in the training set). In this paper, we study LMTC under the zero-shot setting, which does not require any annotated documents with labels and only relies on label surface names and descriptions. To train a classifier that calculates the similarity score between a document and a label, we propose a novel metadata-induced contrastive learning (MICoL) method. Different from previous text-based contrastive learning techniques, MICoL exploits document metadata (e.g., authors, venues, and references of research papers), which are widely available on the Web, to derive similar document-document pairs. Experimental results on two large-scale datasets show that: (1) MICoL significantly outperforms strong zero-shot text classification and contrastive learning baselines; (2) MICoL is on par with the state-of-the-art supervised metadata-aware LMTC method trained on 10K-200K labeled documents; and (3) MICoL tends to predict more infrequent labels than supervised methods, thus alleviates the deteriorated performance on long-tailed labels.
Universal Representation Learning of Knowledge Bases by Jointly Embedding Instances and Ontological Concepts
Mar 15, 2021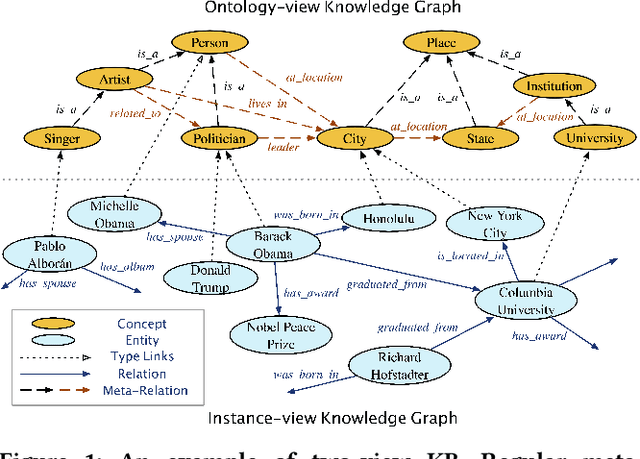
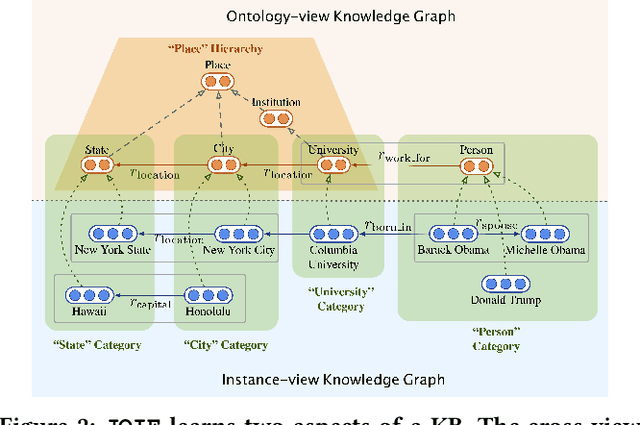
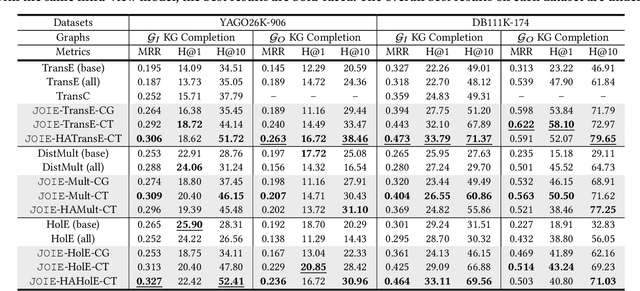
Abstract:Many large-scale knowledge bases simultaneously represent two views of knowledge graphs (KGs): an ontology view for abstract and commonsense concepts, and an instance view for specific entities that are instantiated from ontological concepts. Existing KG embedding models, however, merely focus on representing one of the two views alone. In this paper, we propose a novel two-view KG embedding model, JOIE, with the goal to produce better knowledge embedding and enable new applications that rely on multi-view knowledge. JOIE employs both cross-view and intra-view modeling that learn on multiple facets of the knowledge base. The cross-view association model is learned to bridge the embeddings of ontological concepts and their corresponding instance-view entities. The intra-view models are trained to capture the structured knowledge of instance and ontology views in separate embedding spaces, with a hierarchy-aware encoding technique enabled for ontologies with hierarchies. We explore multiple representation techniques for the two model components and investigate with nine variants of JOIE. Our model is trained on large-scale knowledge bases that consist of massive instances and their corresponding ontological concepts connected via a (small) set of cross-view links. Experimental results on public datasets show that the best variant of JOIE significantly outperforms previous models on instance-view triple prediction task as well as ontology population on ontologyview KG. In addition, our model successfully extends the use of KG embeddings to entity typing with promising performance.
Bio-JOIE: Joint Representation Learning of Biological Knowledge Bases
Mar 07, 2021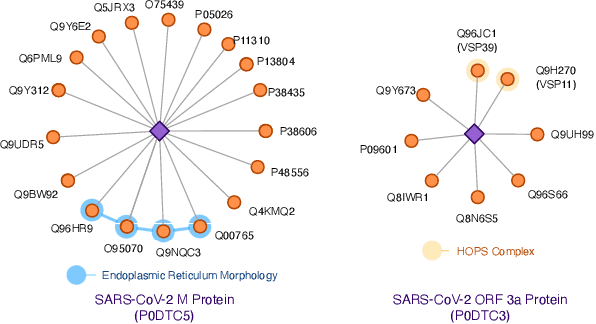
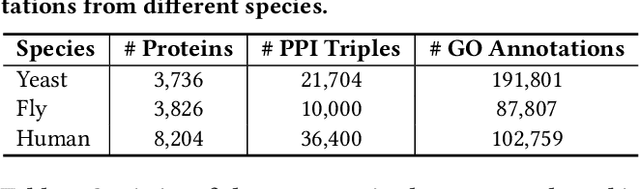
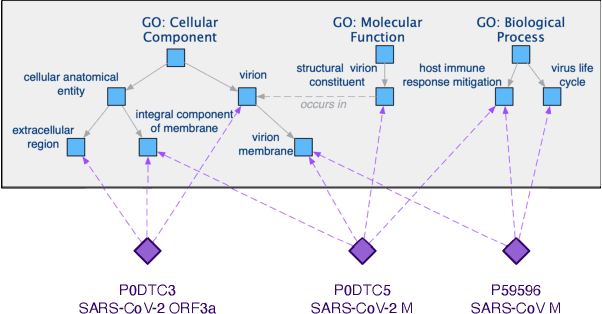
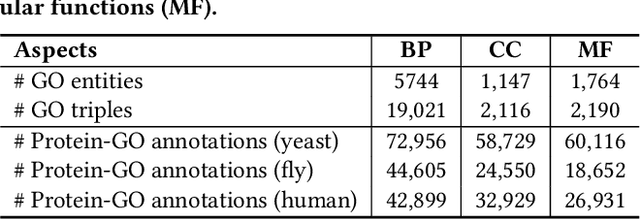
Abstract:The widespread of Coronavirus has led to a worldwide pandemic with a high mortality rate. Currently, the knowledge accumulated from different studies about this virus is very limited. Leveraging a wide-range of biological knowledge, such as gene ontology and protein-protein interaction (PPI) networks from other closely related species presents a vital approach to infer the molecular impact of a new species. In this paper, we propose the transferred multi-relational embedding model Bio-JOIE to capture the knowledge of gene ontology and PPI networks, which demonstrates superb capability in modeling the SARS-CoV-2-human protein interactions. Bio-JOIE jointly trains two model components. The knowledge model encodes the relational facts from the protein and GO domains into separated embedding spaces, using a hierarchy-aware encoding technique employed for the GO terms. On top of that, the transfer model learns a non-linear transformation to transfer the knowledge of PPIs and gene ontology annotations across their embedding spaces. By leveraging only structured knowledge, Bio-JOIE significantly outperforms existing state-of-the-art methods in PPI type prediction on multiple species. Furthermore, we also demonstrate the potential of leveraging the learned representations on clustering proteins with enzymatic function into enzyme commission families. Finally, we show that Bio-JOIE can accurately identify PPIs between the SARS-CoV-2 proteins and human proteins, providing valuable insights for advancing research on this new disease.
* ACM BCB 2020, Best Student Paper
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge