Jongseong Jang
EXAONE Path 2.5: Pathology Foundation Model with Multi-Omics Alignment
Dec 16, 2025Abstract:Cancer progression arises from interactions across multiple biological layers, especially beyond morphological and across molecular layers that remain invisible to image-only models. To capture this broader biological landscape, we present EXAONE Path 2.5, a pathology foundation model that jointly models histologic, genomic, epigenetic and transcriptomic modalities, producing an integrated patient representation that reflects tumor biology more comprehensively. Our approach incorporates three key components: (1) multimodal SigLIP loss enabling all-pairwise contrastive learning across heterogeneous modalities, (2) a fragment-aware rotary positional encoding (F-RoPE) module that preserves spatial structure and tissue-fragment topology in WSI, and (3) domain-specialized internal foundation models for both WSI and RNA-seq to provide biologically grounded embeddings for robust multimodal alignment. We evaluate EXAONE Path 2.5 against six leading pathology foundation models across two complementary benchmarks: an internal real-world clinical dataset and the Patho-Bench benchmark covering 80 tasks. Our framework demonstrates high data and parameter efficiency, achieving on-par performance with state-of-the-art foundation models on Patho-Bench while exhibiting the highest adaptability in the internal clinical setting. These results highlight the value of biologically informed multimodal design and underscore the potential of integrated genotype-to-phenotype modeling for next-generation precision oncology.
EXAONE Path 2.0: Pathology Foundation Model with End-to-End Supervision
Jul 09, 2025Abstract:In digital pathology, whole-slide images (WSIs) are often difficult to handle due to their gigapixel scale, so most approaches train patch encoders via self-supervised learning (SSL) and then aggregate the patch-level embeddings via multiple instance learning (MIL) or slide encoders for downstream tasks. However, patch-level SSL may overlook complex domain-specific features that are essential for biomarker prediction, such as mutation status and molecular characteristics, as SSL methods rely only on basic augmentations selected for natural image domains on small patch-level area. Moreover, SSL methods remain less data efficient than fully supervised approaches, requiring extensive computational resources and datasets to achieve competitive performance. To address these limitations, we present EXAONE Path 2.0, a pathology foundation model that learns patch-level representations under direct slide-level supervision. Using only 37k WSIs for training, EXAONE Path 2.0 achieves state-of-the-art average performance across 10 biomarker prediction tasks, demonstrating remarkable data efficiency.
ChatEXAONEPath: An Expert-level Multimodal Large Language Model for Histopathology Using Whole Slide Images
Apr 17, 2025Abstract:Recent studies have made significant progress in developing large language models (LLMs) in the medical domain, which can answer expert-level questions and demonstrate the potential to assist clinicians in real-world clinical scenarios. Studies have also witnessed the importance of integrating various modalities with the existing LLMs for a better understanding of complex clinical contexts, which are innately multi-faceted by nature. Although studies have demonstrated the ability of multimodal LLMs in histopathology to answer questions from given images, they lack in understanding of thorough clinical context due to the patch-level data with limited information from public datasets. Thus, developing WSI-level MLLMs is significant in terms of the scalability and applicability of MLLMs in histopathology. In this study, we introduce an expert-level MLLM for histopathology using WSIs, dubbed as ChatEXAONEPath. We present a retrieval-based data generation pipeline using 10,094 pairs of WSIs and histopathology reports from The Cancer Genome Atlas (TCGA). We also showcase an AI-based evaluation protocol for a comprehensive understanding of the medical context from given multimodal information and evaluate generated answers compared to the original histopathology reports. We demonstrate the ability of diagnosing the given histopathology images using ChatEXAONEPath with the acceptance rate of 62.9% from 1,134 pairs of WSIs and reports. Our proposed model can understand pan-cancer WSIs and clinical context from various cancer types. We argue that our proposed model has the potential to assist clinicians by comprehensively understanding complex morphology of WSIs for cancer diagnosis through the integration of multiple modalities.
Pan-cancer gene set discovery via scRNA-seq for optimal deep learning based downstream tasks
Aug 13, 2024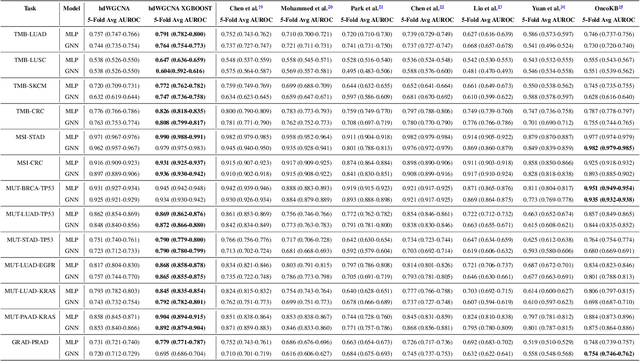
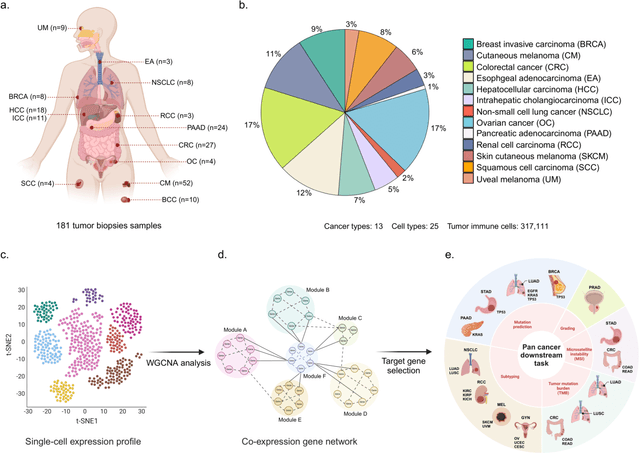
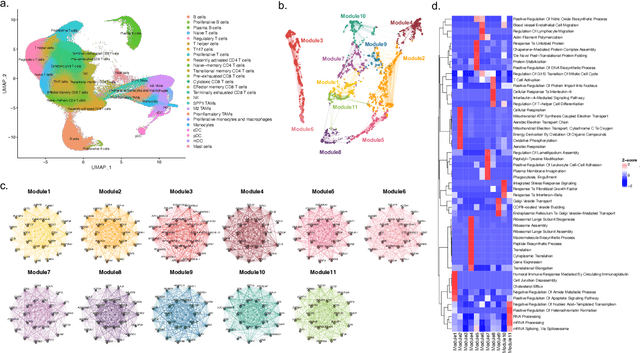
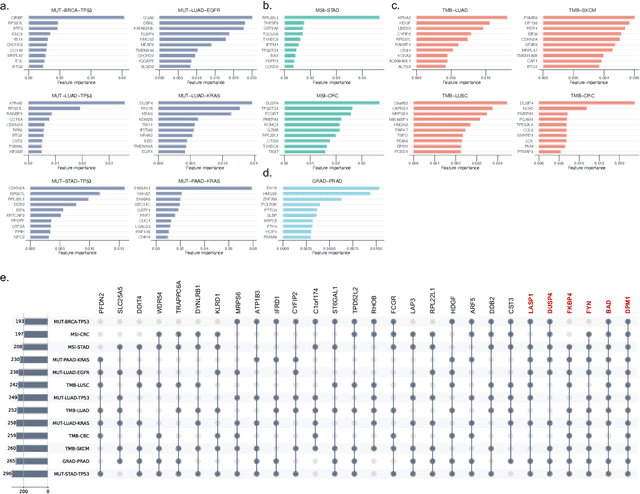
Abstract:The application of machine learning to transcriptomics data has led to significant advances in cancer research. However, the high dimensionality and complexity of RNA sequencing (RNA-seq) data pose significant challenges in pan-cancer studies. This study hypothesizes that gene sets derived from single-cell RNA sequencing (scRNA-seq) data will outperform those selected using bulk RNA-seq in pan-cancer downstream tasks. We analyzed scRNA-seq data from 181 tumor biopsies across 13 cancer types. High-dimensional weighted gene co-expression network analysis (hdWGCNA) was performed to identify relevant gene sets, which were further refined using XGBoost for feature selection. These gene sets were applied to downstream tasks using TCGA pan-cancer RNA-seq data and compared to six reference gene sets and oncogenes from OncoKB evaluated with deep learning models, including multilayer perceptrons (MLPs) and graph neural networks (GNNs). The XGBoost-refined hdWGCNA gene set demonstrated higher performance in most tasks, including tumor mutation burden assessment, microsatellite instability classification, mutation prediction, cancer subtyping, and grading. In particular, genes such as DPM1, BAD, and FKBP4 emerged as important pan-cancer biomarkers, with DPM1 consistently significant across tasks. This study presents a robust approach for feature selection in cancer genomics by integrating scRNA-seq data and advanced analysis techniques, offering a promising avenue for improving predictive accuracy in cancer research.
Enhancing Whole Slide Pathology Foundation Models through Stain Normalization
Aug 05, 2024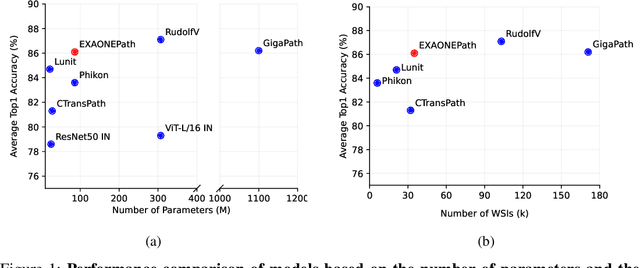
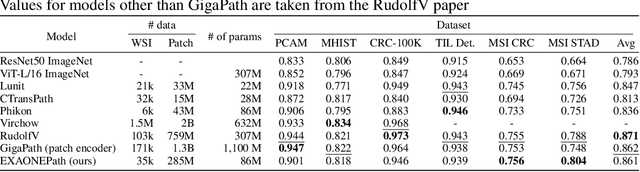
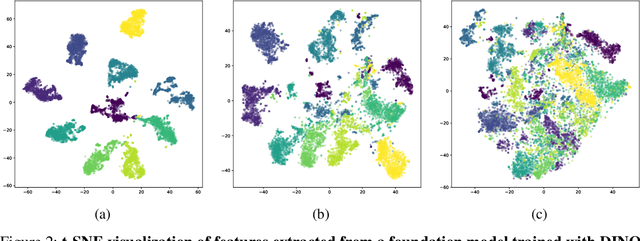
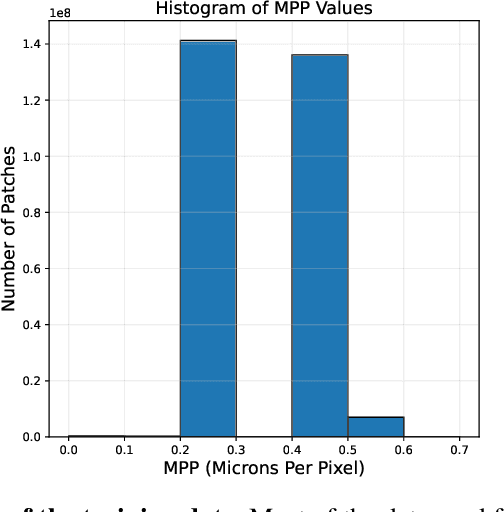
Abstract:Recent advancements in digital pathology have led to the development of numerous foundational models that utilize self-supervised learning on patches extracted from gigapixel whole slide images (WSIs). While this approach leverages vast amounts of unlabeled data, we have discovered a significant issue: features extracted from these self-supervised models tend to cluster by individual WSIs, a phenomenon we term WSI-specific feature collapse. This problem can potentially limit the model's generalization ability and performance on various downstream tasks. To address this issue, we introduce Stain Normalized Pathology Foundational Model, a novel foundational model trained on patches that have undergone stain normalization. Stain normalization helps reduce color variability arising from different laboratories and scanners, enabling the model to learn more consistent features. Stain Normalized Pathology Foundational Model is trained using 285,153,903 patches extracted from a total of 34,795 WSIs, combining data from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) project. Our experiments demonstrate that Stain Normalized Pathology Foundational Model significantly mitigates the feature collapse problem, indicating that the model has learned more generalized features rather than overfitting to individual WSI characteristics. We compared Stain Normalized Pathology Foundational Model with state-of-the-art models across six downstream task datasets, and our results show that Stain Normalized Pathology Foundational Model achieves excellent performance relative to the number of WSIs used and the model's parameter count. This suggests that the application of stain normalization has substantially improved the model's efficiency and generalization capabilities.
Significantly improving zero-shot X-ray pathology classification via fine-tuning pre-trained image-text encoders
Dec 14, 2022Abstract:Deep neural networks have been successfully adopted to diverse domains including pathology classification based on medical images. However, large-scale and high-quality data to train powerful neural networks are rare in the medical domain as the labeling must be done by qualified experts. Researchers recently tackled this problem with some success by taking advantage of models pre-trained on large-scale general domain data. Specifically, researchers took contrastive image-text encoders (e.g., CLIP) and fine-tuned it with chest X-ray images and paired reports to perform zero-shot pathology classification, thus completely removing the need for pathology-annotated images to train a classification model. Existing studies, however, fine-tuned the pre-trained model with the same contrastive learning objective, and failed to exploit the multi-labeled nature of medical image-report pairs. In this paper, we propose a new fine-tuning strategy based on sentence sampling and positive-pair loss relaxation for improving the downstream zero-shot pathology classification performance, which can be applied to any pre-trained contrastive image-text encoders. Our method consistently showed dramatically improved zero-shot pathology classification performance on four different chest X-ray datasets and 3 different pre-trained models (5.77% average AUROC increase). In particular, fine-tuning CLIP with our method showed much comparable or marginally outperformed to board-certified radiologists (0.619 vs 0.625 in F1 score and 0.530 vs 0.544 in MCC) in zero-shot classification of five prominent diseases from the CheXpert dataset.
TransCAM: Transformer Attention-based CAM Refinement for Weakly Supervised Semantic Segmentation
Mar 14, 2022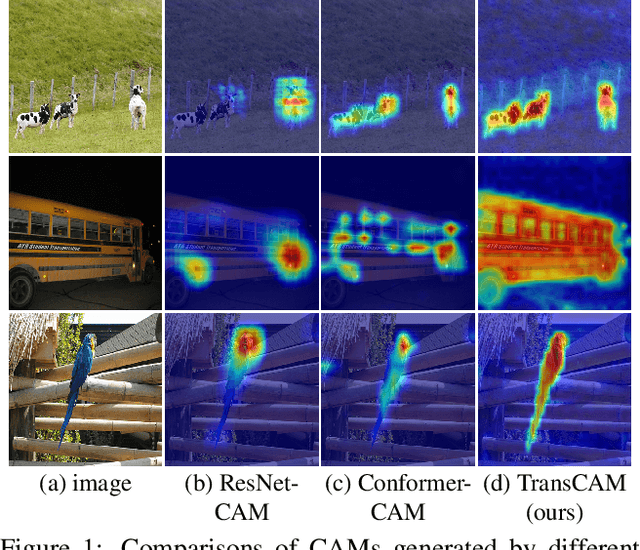
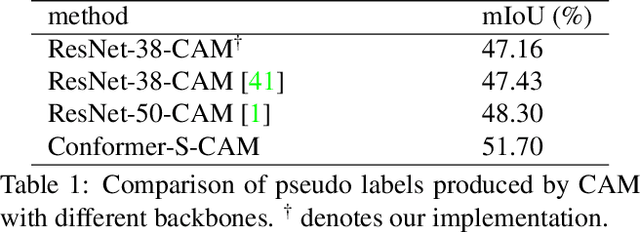
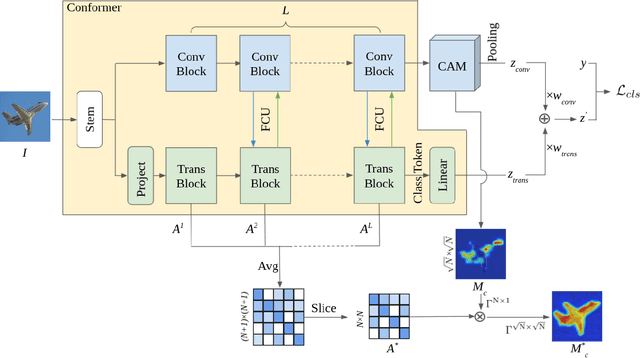

Abstract:Weakly supervised semantic segmentation (WSSS) with only image-level supervision is a challenging task. Most existing methods exploit Class Activation Maps (CAM) to generate pixel-level pseudo labels for supervised training. However, due to the local receptive field of Convolution Neural Networks (CNN), CAM applied to CNNs often suffers from partial activation -- highlighting the most discriminative part instead of the entire object area. In order to capture both local features and global representations, the Conformer has been proposed to combine a visual transformer branch with a CNN branch. In this paper, we propose TransCAM, a Conformer-based solution to WSSS that explicitly leverages the attention weights from the transformer branch of the Conformer to refine the CAM generated from the CNN branch. TransCAM is motivated by our observation that attention weights from shallow transformer blocks are able to capture low-level spatial feature similarities while attention weights from deep transformer blocks capture high-level semantic context. Despite its simplicity, TransCAM achieves a new state-of-the-art performance of 69.3% and 69.6% on the respective PASCAL VOC 2012 validation and test sets, showing the effectiveness of transformer attention-based refinement of CAM for WSSS.
ExCon: Explanation-driven Supervised Contrastive Learning for Image Classification
Dec 10, 2021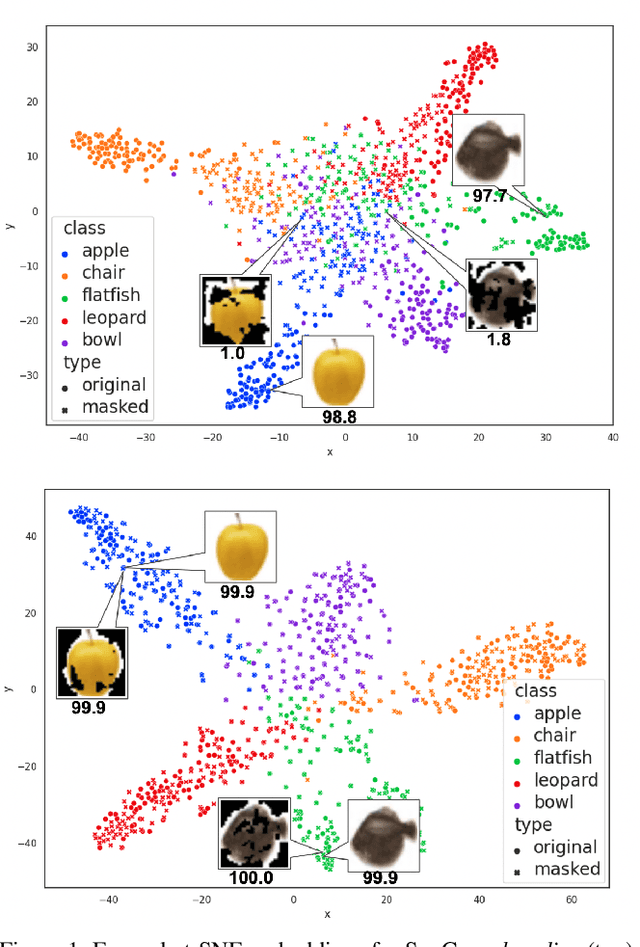
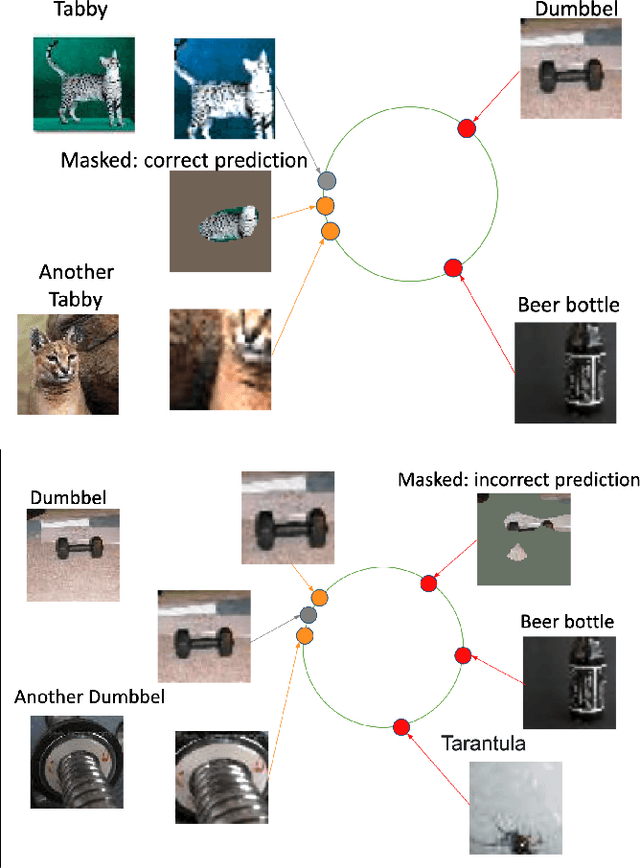
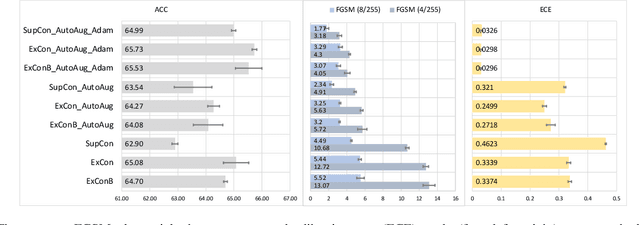
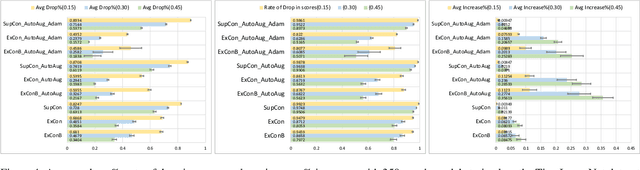
Abstract:Contrastive learning has led to substantial improvements in the quality of learned embedding representations for tasks such as image classification. However, a key drawback of existing contrastive augmentation methods is that they may lead to the modification of the image content which can yield undesired alterations of its semantics. This can affect the performance of the model on downstream tasks. Hence, in this paper, we ask whether we can augment image data in contrastive learning such that the task-relevant semantic content of an image is preserved. For this purpose, we propose to leverage saliency-based explanation methods to create content-preserving masked augmentations for contrastive learning. Our novel explanation-driven supervised contrastive learning (ExCon) methodology critically serves the dual goals of encouraging nearby image embeddings to have similar content and explanation. To quantify the impact of ExCon, we conduct experiments on the CIFAR-100 and the Tiny ImageNet datasets. We demonstrate that ExCon outperforms vanilla supervised contrastive learning in terms of classification, explanation quality, adversarial robustness as well as calibration of probabilistic predictions of the model in the context of distributional shift.
Look at here : Utilizing supervision to attend subtle key regions
Nov 25, 2021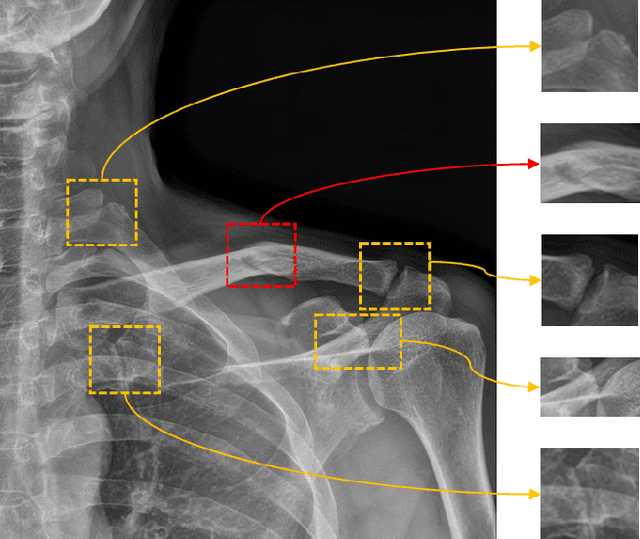
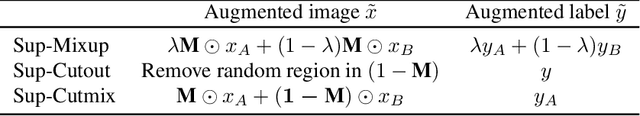
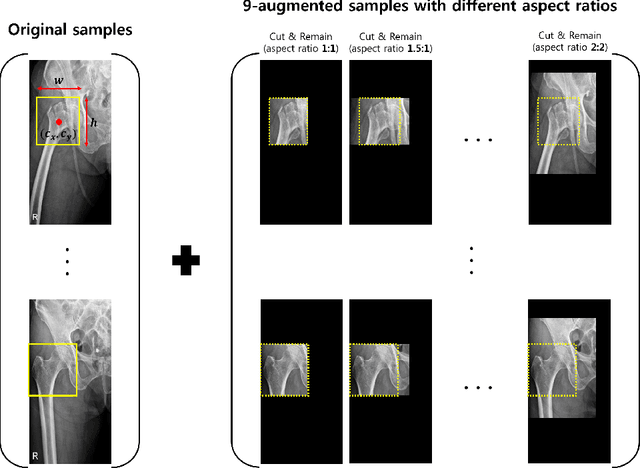
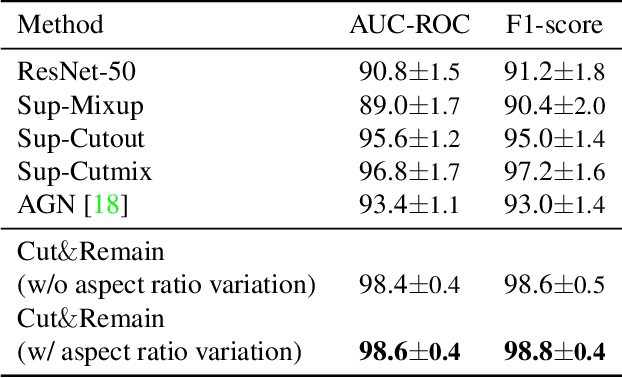
Abstract:Despite the success of deep learning in computer vision, algorithms to recognize subtle and small objects (or regions) is still challenging. For example, recognizing a baseball or a frisbee on a ground scene or a bone fracture in an X-ray image can easily result in overfitting, unless a huge amount of training data is available. To mitigate this problem, we need a way to force a model should identify subtle regions in limited training data. In this paper, we propose a simple but efficient supervised augmentation method called Cut\&Remain. It achieved better performance on various medical image domain (internally sourced- and public dataset) and a natural image domain (MS-COCO$_s$) than other supervised augmentation and the explicit guidance methods. In addition, using the class activation map, we identified that the Cut\&Remain methods drive a model to focus on relevant subtle and small regions efficiently. We also show that the performance monotonically increased along the Cut\&Remain ratio, indicating that a model can be improved even though only limited amount of Cut\&Remain is applied for, so that it allows low supervising (annotation) cost for improvement.
EDDA: Explanation-driven Data Augmentation to Improve Model and Explanation Alignment
Jun 19, 2021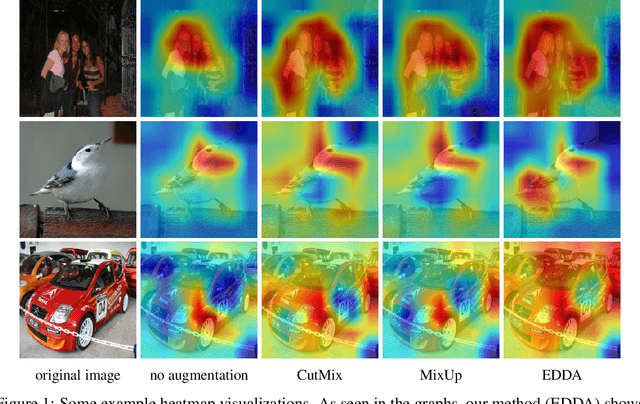
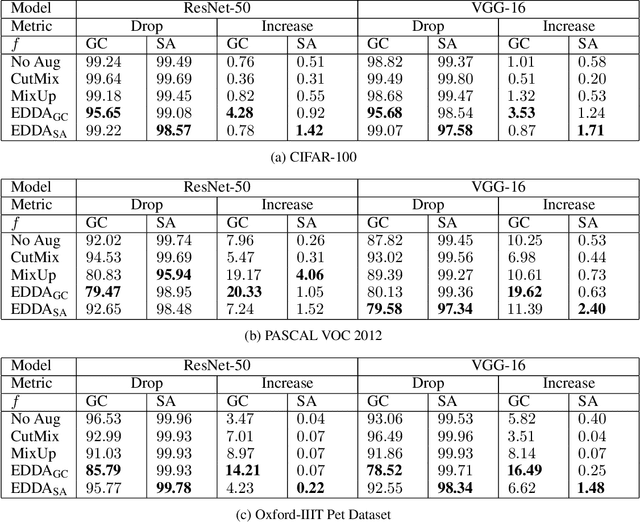
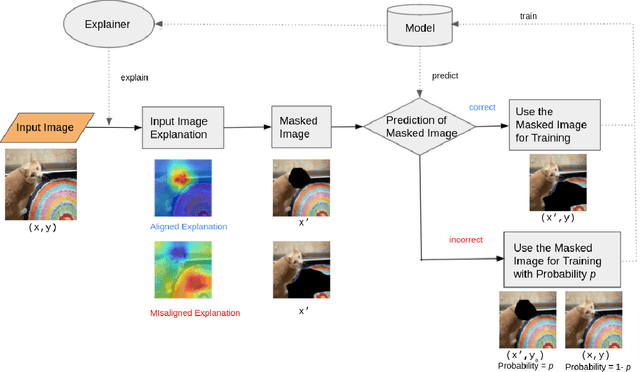
Abstract:Recent years have seen the introduction of a range of methods for post-hoc explainability of image classifier predictions. However, these post-hoc explanations may not always align perfectly with classifier predictions, which poses a significant challenge when attempting to debug models based on such explanations. To this end, we seek a methodology that can improve alignment between model predictions and explanation method that is both agnostic to the model and explanation classes and which does not require ground truth explanations. We achieve this through a novel explanation-driven data augmentation (EDDA) method that augments the training data with occlusions of existing data stemming from model-explanations; this is based on the simple motivating principle that occluding salient regions for the model prediction should decrease the model confidence in the prediction, while occluding non-salient regions should not change the prediction -- if the model and explainer are aligned. To verify that this augmentation method improves model and explainer alignment, we evaluate the methodology on a variety of datasets, image classification models, and explanation methods. We verify in all cases that our explanation-driven data augmentation method improves alignment of the model and explanation in comparison to no data augmentation and non-explanation driven data augmentation methods. In conclusion, this approach provides a novel model- and explainer-agnostic methodology for improving alignment between model predictions and explanations, which we see as a critical step forward for practical deployment and debugging of image classification models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge