Daeun Kyung
ECG-Agent: On-Device Tool-Calling Agent for ECG Multi-Turn Dialogue
Jan 28, 2026Abstract:Recent advances in Multimodal Large Language Models have rapidly expanded to electrocardiograms, focusing on classification, report generation, and single-turn QA tasks. However, these models fall short in real-world scenarios, lacking multi-turn conversational ability, on-device efficiency, and precise understanding of ECG measurements such as the PQRST intervals. To address these limitations, we introduce ECG-Agent, the first LLM-based tool-calling agent for multi-turn ECG dialogue. To facilitate its development and evaluation, we also present ECG-Multi-Turn-Dialogue (ECG-MTD) dataset, a collection of realistic user-assistant multi-turn dialogues for diverse ECG lead configurations. We develop ECG-Agents in various sizes, from on-device capable to larger agents. Experimental results show that ECG-Agents outperform baseline ECG-LLMs in response accuracy. Furthermore, on-device agents achieve comparable performance to larger agents in various evaluations that assess response accuracy, tool-calling ability, and hallucinations, demonstrating their viability for real-world applications.
PatientSim: A Persona-Driven Simulator for Realistic Doctor-Patient Interactions
May 23, 2025
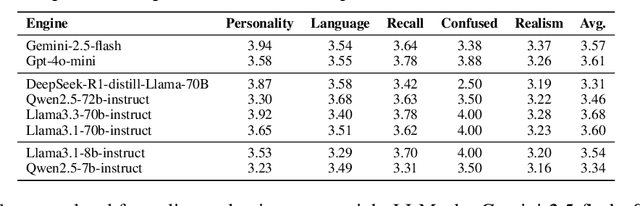
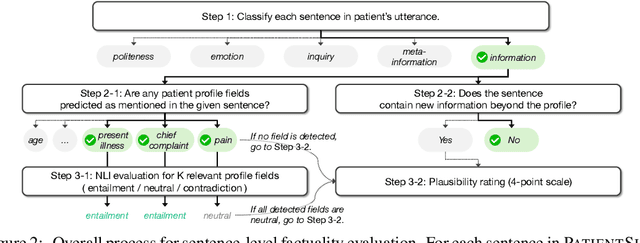
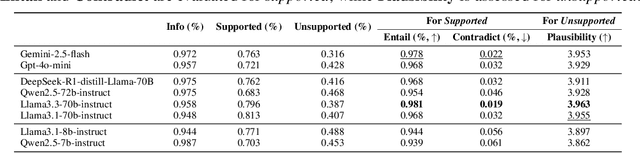
Abstract:Doctor-patient consultations require multi-turn, context-aware communication tailored to diverse patient personas. Training or evaluating doctor LLMs in such settings requires realistic patient interaction systems. However, existing simulators often fail to reflect the full range of personas seen in clinical practice. To address this, we introduce PatientSim, a patient simulator that generates realistic and diverse patient personas for clinical scenarios, grounded in medical expertise. PatientSim operates using: 1) clinical profiles, including symptoms and medical history, derived from real-world data in the MIMIC-ED and MIMIC-IV datasets, and 2) personas defined by four axes: personality, language proficiency, medical history recall level, and cognitive confusion level, resulting in 37 unique combinations. We evaluated eight LLMs for factual accuracy and persona consistency. The top-performing open-source model, Llama 3.3, was validated by four clinicians to confirm the robustness of our framework. As an open-source, customizable platform, PatientSim provides a reproducible and scalable solution that can be customized for specific training needs. Offering a privacy-compliant environment, it serves as a robust testbed for evaluating medical dialogue systems across diverse patient presentations and shows promise as an educational tool for healthcare.
Towards Predicting Temporal Changes in a Patient's Chest X-ray Images based on Electronic Health Records
Sep 11, 2024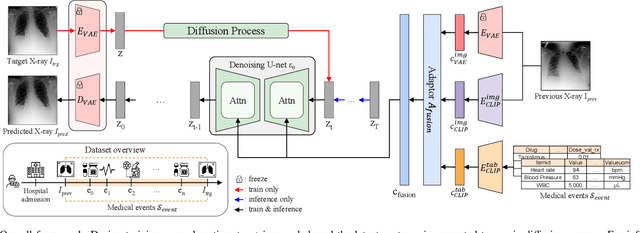
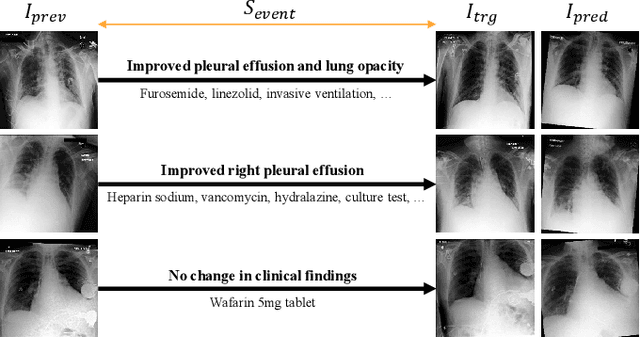
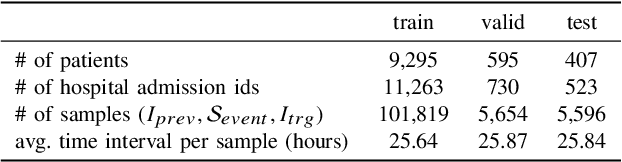
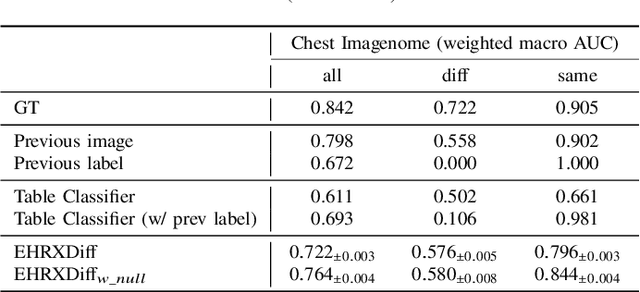
Abstract:Chest X-ray imaging (CXR) is an important diagnostic tool used in hospitals to assess patient conditions and monitor changes over time. Generative models, specifically diffusion-based models, have shown promise in generating realistic synthetic X-rays. However, these models mainly focus on conditional generation using single-time-point data, i.e., typically CXRs taken at a specific time with their corresponding reports, limiting their clinical utility, particularly for capturing temporal changes. To address this limitation, we propose a novel framework, EHRXDiff, which predicts future CXR images by integrating previous CXRs with subsequent medical events, e.g., prescriptions, lab measures, etc. Our framework dynamically tracks and predicts disease progression based on a latent diffusion model, conditioned on the previous CXR image and a history of medical events. We comprehensively evaluate the performance of our framework across three key aspects, including clinical consistency, demographic consistency, and visual realism. We demonstrate that our framework generates high-quality, realistic future images that capture potential temporal changes, suggesting its potential for further development as a clinical simulation tool. This could offer valuable insights for patient monitoring and treatment planning in the medical field.
EHRCon: Dataset for Checking Consistency between Unstructured Notes and Structured Tables in Electronic Health Records
Jun 24, 2024



Abstract:Electronic Health Records (EHRs) are integral for storing comprehensive patient medical records, combining structured data (e.g., medications) with detailed clinical notes (e.g., physician notes). These elements are essential for straightforward data retrieval and provide deep, contextual insights into patient care. However, they often suffer from discrepancies due to unintuitive EHR system designs and human errors, posing serious risks to patient safety. To address this, we developed EHRCon, a new dataset and task specifically designed to ensure data consistency between structured tables and unstructured notes in EHRs. EHRCon was crafted in collaboration with healthcare professionals using the MIMIC-III EHR dataset, and includes manual annotations of 3,943 entities across 105 clinical notes checked against database entries for consistency. EHRCon has two versions, one using the original MIMIC-III schema, and another using the OMOP CDM schema, in order to increase its applicability and generalizability. Furthermore, leveraging the capabilities of large language models, we introduce CheckEHR, a novel framework for verifying the consistency between clinical notes and database tables. CheckEHR utilizes an eight-stage process and shows promising results in both few-shot and zero-shot settings. The code is available at https://github.com/dustn1259/EHRCon.
DialSim: A Real-Time Simulator for Evaluating Long-Term Dialogue Understanding of Conversational Agents
Jun 19, 2024



Abstract:Recent advancements in Large Language Models (LLMs) have significantly enhanced the capabilities of conversational agents, making them applicable to various fields (e.g., education). Despite their progress, the evaluation of the agents often overlooks the complexities of real-world conversations, such as real-time interactions, multi-party dialogues, and extended contextual dependencies. To bridge this gap, we introduce DialSim, a real-time dialogue simulator. In this simulator, an agent is assigned the role of a character from popular TV shows, requiring it to respond to spontaneous questions using past dialogue information and to distinguish between known and unknown information. Key features of DialSim include evaluating the agent's ability to respond within a reasonable time limit, handling long-term multi-party dialogues, and managing adversarial settings (e.g., swap character names) to challenge the agent's reliance on pre-trained knowledge. We utilized this simulator to evaluate the latest conversational agents and analyze their limitations. Our experiments highlight both the strengths and weaknesses of these agents, providing valuable insights for future improvements in the field of conversational AI. DialSim is available at https://github.com/jiho283/Simulator.
EHRXQA: A Multi-Modal Question Answering Dataset for Electronic Health Records with Chest X-ray Images
Oct 28, 2023



Abstract:Electronic Health Records (EHRs), which contain patients' medical histories in various multi-modal formats, often overlook the potential for joint reasoning across imaging and table modalities underexplored in current EHR Question Answering (QA) systems. In this paper, we introduce EHRXQA, a novel multi-modal question answering dataset combining structured EHRs and chest X-ray images. To develop our dataset, we first construct two uni-modal resources: 1) The MIMIC- CXR-VQA dataset, our newly created medical visual question answering (VQA) benchmark, specifically designed to augment the imaging modality in EHR QA, and 2) EHRSQL (MIMIC-IV), a refashioned version of a previously established table-based EHR QA dataset. By integrating these two uni-modal resources, we successfully construct a multi-modal EHR QA dataset that necessitates both uni-modal and cross-modal reasoning. To address the unique challenges of multi-modal questions within EHRs, we propose a NeuralSQL-based strategy equipped with an external VQA API. This pioneering endeavor enhances engagement with multi-modal EHR sources and we believe that our dataset can catalyze advances in real-world medical scenarios such as clinical decision-making and research. EHRXQA is available at https://github.com/baeseongsu/ehrxqa.
Perspective Projection-Based 3D CT Reconstruction from Biplanar X-rays
Mar 09, 2023



Abstract:X-ray computed tomography (CT) is one of the most common imaging techniques used to diagnose various diseases in the medical field. Its high contrast sensitivity and spatial resolution allow the physician to observe details of body parts such as bones, soft tissue, blood vessels, etc. As it involves potentially harmful radiation exposure to patients and surgeons, however, reconstructing 3D CT volume from perpendicular 2D X-ray images is considered a promising alternative, thanks to its lower radiation risk and better accessibility. This is highly challenging though, since it requires reconstruction of 3D anatomical information from 2D images with limited views, where all the information is overlapped. In this paper, we propose PerX2CT, a novel CT reconstruction framework from X-ray that reflects the perspective projection scheme. Our proposed method provides a different combination of features for each coordinate which implicitly allows the model to obtain information about the 3D location. We reveal the potential to reconstruct the selected part of CT with high resolution by properly using the coordinate-wise local and global features. Our approach shows potential for use in clinical applications with low computational complexity and fast inference time, demonstrating superior performance than baselines in multiple evaluation metrics.
Significantly improving zero-shot X-ray pathology classification via fine-tuning pre-trained image-text encoders
Dec 14, 2022Abstract:Deep neural networks have been successfully adopted to diverse domains including pathology classification based on medical images. However, large-scale and high-quality data to train powerful neural networks are rare in the medical domain as the labeling must be done by qualified experts. Researchers recently tackled this problem with some success by taking advantage of models pre-trained on large-scale general domain data. Specifically, researchers took contrastive image-text encoders (e.g., CLIP) and fine-tuned it with chest X-ray images and paired reports to perform zero-shot pathology classification, thus completely removing the need for pathology-annotated images to train a classification model. Existing studies, however, fine-tuned the pre-trained model with the same contrastive learning objective, and failed to exploit the multi-labeled nature of medical image-report pairs. In this paper, we propose a new fine-tuning strategy based on sentence sampling and positive-pair loss relaxation for improving the downstream zero-shot pathology classification performance, which can be applied to any pre-trained contrastive image-text encoders. Our method consistently showed dramatically improved zero-shot pathology classification performance on four different chest X-ray datasets and 3 different pre-trained models (5.77% average AUROC increase). In particular, fine-tuning CLIP with our method showed much comparable or marginally outperformed to board-certified radiologists (0.619 vs 0.625 in F1 score and 0.530 vs 0.544 in MCC) in zero-shot classification of five prominent diseases from the CheXpert dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge