Sunjun Kweon
Overview of the EHRSQL 2024 Shared Task on Reliable Text-to-SQL Modeling on Electronic Health Records
May 04, 2024Abstract:Electronic Health Records (EHRs) are relational databases that store the entire medical histories of patients within hospitals. They record numerous aspects of patients' medical care, from hospital admission and diagnosis to treatment and discharge. While EHRs are vital sources of clinical data, exploring them beyond a predefined set of queries requires skills in query languages like SQL. To make information retrieval more accessible, one strategy is to build a question-answering system, possibly leveraging text-to-SQL models that can automatically translate natural language questions into corresponding SQL queries and use these queries to retrieve the answers. The EHRSQL 2024 shared task aims to advance and promote research in developing a question-answering system for EHRs using text-to-SQL modeling, capable of reliably providing requested answers to various healthcare professionals to improve their clinical work processes and satisfy their needs. Among more than 100 participants who applied to the shared task, eight teams completed the entire shared task processes and demonstrated a wide range of methods to effectively solve this task. In this paper, we describe the task of reliable text-to-SQL modeling, the dataset, and the methods and results of the participants. We hope this shared task will spur further research and insights into developing reliable question-answering systems for EHRs.
KorMedMCQA: Multi-Choice Question Answering Benchmark for Korean Healthcare Professional Licensing Examinations
Mar 05, 2024Abstract:We introduce KorMedMCQA, the first Korean multiple-choice question answering (MCQA) benchmark derived from Korean healthcare professional licensing examinations, covering from the year 2012 to year 2023. This dataset consists of a selection of questions from the license examinations for doctors, nurses, and pharmacists, featuring a diverse array of subjects. We conduct baseline experiments on various large language models, including proprietary/open-source, multilingual/Korean-additional pretrained, and clinical context pretrained models, highlighting the potential for further enhancements. We make our data publicly available on HuggingFace (https://huggingface.co/datasets/sean0042/KorMedMCQA) and provide a evaluation script via LM-Harness, inviting further exploration and advancement in Korean healthcare environments.
EHRNoteQA: A Patient-Specific Question Answering Benchmark for Evaluating Large Language Models in Clinical Settings
Feb 27, 2024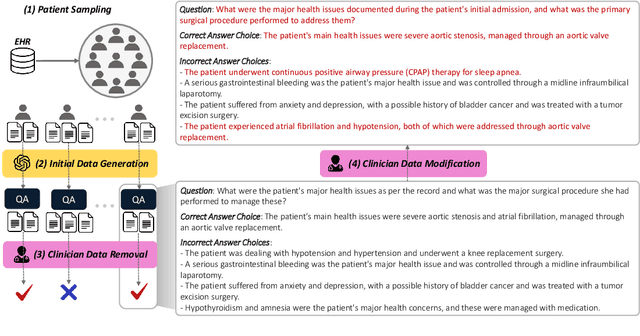
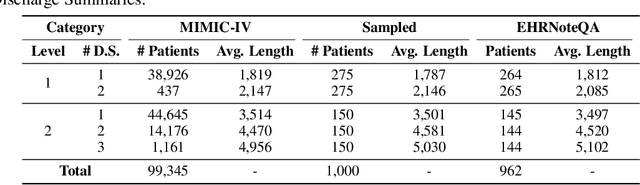


Abstract:This study introduces EHRNoteQA, a novel patient-specific question answering benchmark tailored for evaluating Large Language Models (LLMs) in clinical environments. Based on MIMIC-IV Electronic Health Record (EHR), a team of three medical professionals has curated the dataset comprising 962 unique questions, each linked to a specific patient's EHR clinical notes. What makes EHRNoteQA distinct from existing EHR-based benchmarks is as follows: Firstly, it is the first dataset to adopt a multi-choice question answering format, a design choice that effectively evaluates LLMs with reliable scores in the context of automatic evaluation, compared to other formats. Secondly, it requires an analysis of multiple clinical notes to answer a single question, reflecting the complex nature of real-world clinical decision-making where clinicians review extensive records of patient histories. Our comprehensive evaluation on various large language models showed that their scores on EHRNoteQA correlate more closely with their performance in addressing real-world medical questions evaluated by clinicians than their scores from other LLM benchmarks. This underscores the significance of EHRNoteQA in evaluating LLMs for medical applications and highlights its crucial role in facilitating the integration of LLMs into healthcare systems. The dataset will be made available to the public under PhysioNet credential access, promoting further research in this vital field.
EHRXQA: A Multi-Modal Question Answering Dataset for Electronic Health Records with Chest X-ray Images
Oct 28, 2023



Abstract:Electronic Health Records (EHRs), which contain patients' medical histories in various multi-modal formats, often overlook the potential for joint reasoning across imaging and table modalities underexplored in current EHR Question Answering (QA) systems. In this paper, we introduce EHRXQA, a novel multi-modal question answering dataset combining structured EHRs and chest X-ray images. To develop our dataset, we first construct two uni-modal resources: 1) The MIMIC- CXR-VQA dataset, our newly created medical visual question answering (VQA) benchmark, specifically designed to augment the imaging modality in EHR QA, and 2) EHRSQL (MIMIC-IV), a refashioned version of a previously established table-based EHR QA dataset. By integrating these two uni-modal resources, we successfully construct a multi-modal EHR QA dataset that necessitates both uni-modal and cross-modal reasoning. To address the unique challenges of multi-modal questions within EHRs, we propose a NeuralSQL-based strategy equipped with an external VQA API. This pioneering endeavor enhances engagement with multi-modal EHR sources and we believe that our dataset can catalyze advances in real-world medical scenarios such as clinical decision-making and research. EHRXQA is available at https://github.com/baeseongsu/ehrxqa.
Publicly Shareable Clinical Large Language Model Built on Synthetic Clinical Notes
Sep 06, 2023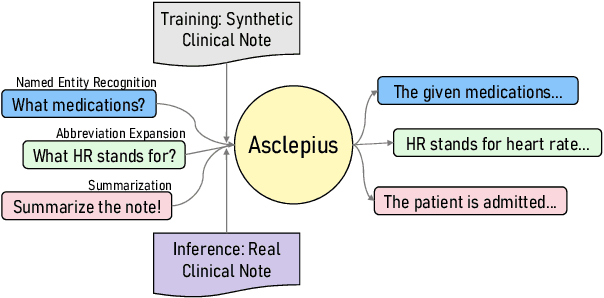
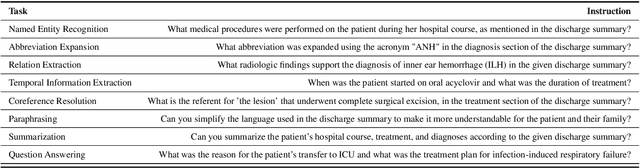
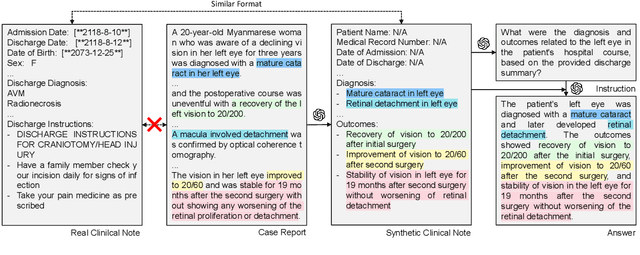
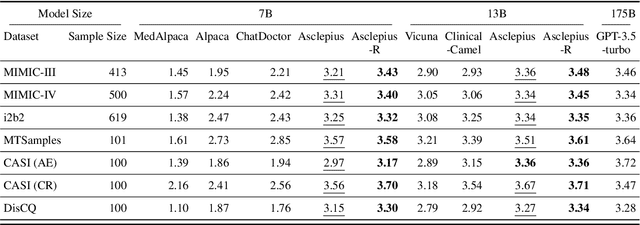
Abstract:The development of large language models tailored for handling patients' clinical notes is often hindered by the limited accessibility and usability of these notes due to strict privacy regulations. To address these challenges, we first create synthetic large-scale clinical notes using publicly available case reports extracted from biomedical literature. We then use these synthetic notes to train our specialized clinical large language model, Asclepius. While Asclepius is trained on synthetic data, we assess its potential performance in real-world applications by evaluating it using real clinical notes. We benchmark Asclepius against several other large language models, including GPT-3.5-turbo and other open-source alternatives. To further validate our approach using synthetic notes, we also compare Asclepius with its variants trained on real clinical notes. Our findings convincingly demonstrate that synthetic clinical notes can serve as viable substitutes for real ones when constructing high-performing clinical language models. This conclusion is supported by detailed evaluations conducted by both GPT-4 and medical professionals. All resources including weights, codes, and data used in the development of Asclepius are made publicly accessible for future research.
Open-WikiTable: Dataset for Open Domain Question Answering with Complex Reasoning over Table
May 12, 2023
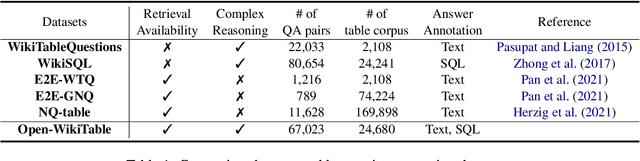


Abstract:Despite recent interest in open domain question answering (ODQA) over tables, many studies still rely on datasets that are not truly optimal for the task with respect to utilizing structural nature of table. These datasets assume answers reside as a single cell value and do not necessitate exploring over multiple cells such as aggregation, comparison, and sorting. Thus, we release Open-WikiTable, the first ODQA dataset that requires complex reasoning over tables. Open-WikiTable is built upon WikiSQL and WikiTableQuestions to be applicable in the open-domain setting. As each question is coupled with both textual answers and SQL queries, Open-WikiTable opens up a wide range of possibilities for future research, as both reader and parser methods can be applied. The dataset and code are publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge