Gyubok Lee
EHRCon: Dataset for Checking Consistency between Unstructured Notes and Structured Tables in Electronic Health Records
Jun 24, 2024



Abstract:Electronic Health Records (EHRs) are integral for storing comprehensive patient medical records, combining structured data (e.g., medications) with detailed clinical notes (e.g., physician notes). These elements are essential for straightforward data retrieval and provide deep, contextual insights into patient care. However, they often suffer from discrepancies due to unintuitive EHR system designs and human errors, posing serious risks to patient safety. To address this, we developed EHRCon, a new dataset and task specifically designed to ensure data consistency between structured tables and unstructured notes in EHRs. EHRCon was crafted in collaboration with healthcare professionals using the MIMIC-III EHR dataset, and includes manual annotations of 3,943 entities across 105 clinical notes checked against database entries for consistency. EHRCon has two versions, one using the original MIMIC-III schema, and another using the OMOP CDM schema, in order to increase its applicability and generalizability. Furthermore, leveraging the capabilities of large language models, we introduce CheckEHR, a novel framework for verifying the consistency between clinical notes and database tables. CheckEHR utilizes an eight-stage process and shows promising results in both few-shot and zero-shot settings. The code is available at https://github.com/dustn1259/EHRCon.
EHR-SeqSQL : A Sequential Text-to-SQL Dataset For Interactively Exploring Electronic Health Records
May 23, 2024Abstract:In this paper, we introduce EHR-SeqSQL, a novel sequential text-to-SQL dataset for Electronic Health Record (EHR) databases. EHR-SeqSQL is designed to address critical yet underexplored aspects in text-to-SQL parsing: interactivity, compositionality, and efficiency. To the best of our knowledge, EHR-SeqSQL is not only the largest but also the first medical text-to-SQL dataset benchmark to include sequential and contextual questions. We provide a data split and the new test set designed to assess compositional generalization ability. Our experiments demonstrate the superiority of a multi-turn approach over a single-turn approach in learning compositionality. Additionally, our dataset integrates specially crafted tokens into SQL queries to improve execution efficiency. With EHR-SeqSQL, we aim to bridge the gap between practical needs and academic research in the text-to-SQL domain.
Overview of the EHRSQL 2024 Shared Task on Reliable Text-to-SQL Modeling on Electronic Health Records
May 04, 2024Abstract:Electronic Health Records (EHRs) are relational databases that store the entire medical histories of patients within hospitals. They record numerous aspects of patients' medical care, from hospital admission and diagnosis to treatment and discharge. While EHRs are vital sources of clinical data, exploring them beyond a predefined set of queries requires skills in query languages like SQL. To make information retrieval more accessible, one strategy is to build a question-answering system, possibly leveraging text-to-SQL models that can automatically translate natural language questions into corresponding SQL queries and use these queries to retrieve the answers. The EHRSQL 2024 shared task aims to advance and promote research in developing a question-answering system for EHRs using text-to-SQL modeling, capable of reliably providing requested answers to various healthcare professionals to improve their clinical work processes and satisfy their needs. Among more than 100 participants who applied to the shared task, eight teams completed the entire shared task processes and demonstrated a wide range of methods to effectively solve this task. In this paper, we describe the task of reliable text-to-SQL modeling, the dataset, and the methods and results of the participants. We hope this shared task will spur further research and insights into developing reliable question-answering systems for EHRs.
Towards Unbiased Evaluation of Detecting Unanswerable Questions in EHRSQL
Apr 29, 2024



Abstract:Incorporating unanswerable questions into EHR QA systems is crucial for testing the trustworthiness of a system, as providing non-existent responses can mislead doctors in their diagnoses. The EHRSQL dataset stands out as a promising benchmark because it is the only dataset that incorporates unanswerable questions in the EHR QA system alongside practical questions. However, in this work, we identify a data bias in these unanswerable questions; they can often be discerned simply by filtering with specific N-gram patterns. Such biases jeopardize the authenticity and reliability of QA system evaluations. To tackle this problem, we propose a simple debiasing method of adjusting the split between the validation and test sets to neutralize the undue influence of N-gram filtering. By experimenting on the MIMIC-III dataset, we demonstrate both the existing data bias in EHRSQL and the effectiveness of our data split strategy in mitigating this bias.
TrustSQL: A Reliability Benchmark for Text-to-SQL Models with Diverse Unanswerable Questions
Mar 23, 2024Abstract:Recent advances in large language models (LLMs) have led to significant improvements in translating natural language questions into SQL queries. While achieving high accuracy in SQL generation is crucial, little is known about the extent to which these text-to-SQL models can reliably handle diverse types of questions encountered during real-world deployment, including unanswerable ones. To explore this aspect, we present TrustSQL, a new benchmark designed to assess the reliability of text-to-SQL models in both single-database and cross-database settings. The benchmark tasks models with providing one of two outcomes: 1) SQL prediction; or 2) abstention from making a prediction, either when there is a potential error in the generated SQL or when faced with unanswerable questions. For model evaluation, we explore various modeling approaches specifically designed for this task. These include: 1) optimizing separate models for answerability detection, SQL generation, and error detection, which are then integrated into a single pipeline; and 2) developing a unified approach that optimizes a single model to address the proposed task. Experimental results using our new reliability score show that addressing this challenge involves many different areas of research and opens new avenues for model development. Nonetheless, none of the methods surpass the reliability performance of the naive baseline, which abstains from answering all questions.
EHRXQA: A Multi-Modal Question Answering Dataset for Electronic Health Records with Chest X-ray Images
Oct 28, 2023



Abstract:Electronic Health Records (EHRs), which contain patients' medical histories in various multi-modal formats, often overlook the potential for joint reasoning across imaging and table modalities underexplored in current EHR Question Answering (QA) systems. In this paper, we introduce EHRXQA, a novel multi-modal question answering dataset combining structured EHRs and chest X-ray images. To develop our dataset, we first construct two uni-modal resources: 1) The MIMIC- CXR-VQA dataset, our newly created medical visual question answering (VQA) benchmark, specifically designed to augment the imaging modality in EHR QA, and 2) EHRSQL (MIMIC-IV), a refashioned version of a previously established table-based EHR QA dataset. By integrating these two uni-modal resources, we successfully construct a multi-modal EHR QA dataset that necessitates both uni-modal and cross-modal reasoning. To address the unique challenges of multi-modal questions within EHRs, we propose a NeuralSQL-based strategy equipped with an external VQA API. This pioneering endeavor enhances engagement with multi-modal EHR sources and we believe that our dataset can catalyze advances in real-world medical scenarios such as clinical decision-making and research. EHRXQA is available at https://github.com/baeseongsu/ehrxqa.
Publicly Shareable Clinical Large Language Model Built on Synthetic Clinical Notes
Sep 06, 2023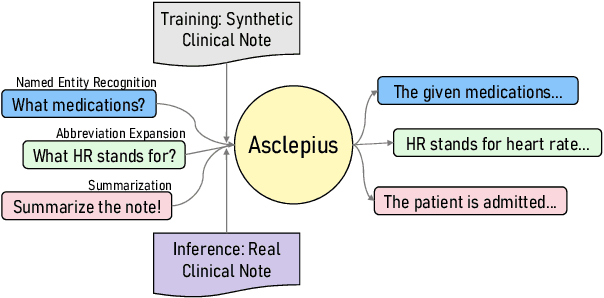
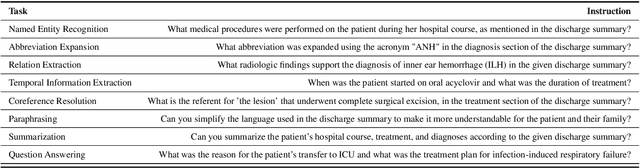
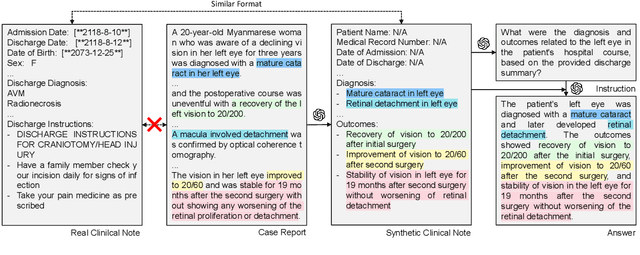
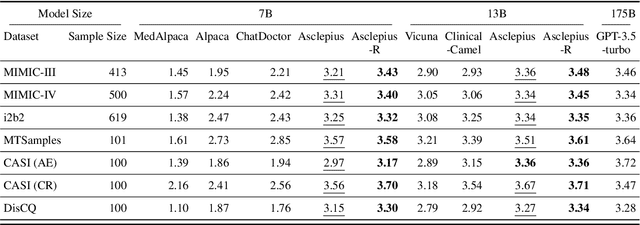
Abstract:The development of large language models tailored for handling patients' clinical notes is often hindered by the limited accessibility and usability of these notes due to strict privacy regulations. To address these challenges, we first create synthetic large-scale clinical notes using publicly available case reports extracted from biomedical literature. We then use these synthetic notes to train our specialized clinical large language model, Asclepius. While Asclepius is trained on synthetic data, we assess its potential performance in real-world applications by evaluating it using real clinical notes. We benchmark Asclepius against several other large language models, including GPT-3.5-turbo and other open-source alternatives. To further validate our approach using synthetic notes, we also compare Asclepius with its variants trained on real clinical notes. Our findings convincingly demonstrate that synthetic clinical notes can serve as viable substitutes for real ones when constructing high-performing clinical language models. This conclusion is supported by detailed evaluations conducted by both GPT-4 and medical professionals. All resources including weights, codes, and data used in the development of Asclepius are made publicly accessible for future research.
ECG-QA: A Comprehensive Question Answering Dataset Combined With Electrocardiogram
Jun 21, 2023



Abstract:Question answering (QA) in the field of healthcare has received much attention due to significant advancements in natural language processing. However, existing healthcare QA datasets primarily focus on medical images, clinical notes, or structured electronic health record tables. This leaves the vast potential of combining electrocardiogram (ECG) data with these systems largely untapped. To address this gap, we present ECG-QA, the first QA dataset specifically designed for ECG analysis. The dataset comprises a total of 70 question templates that cover a wide range of clinically relevant ECG topics, each validated by an ECG expert to ensure their clinical utility. As a result, our dataset includes diverse ECG interpretation questions, including those that require a comparative analysis of two different ECGs. In addition, we have conducted numerous experiments to provide valuable insights for future research directions. We believe that ECG-QA will serve as a valuable resource for the development of intelligent QA systems capable of assisting clinicians in ECG interpretations.
EHRSQL: A Practical Text-to-SQL Benchmark for Electronic Health Records
Jan 16, 2023



Abstract:We present a new text-to-SQL dataset for electronic health records (EHRs). The utterances were collected from 222 hospital staff, including physicians, nurses, insurance review and health records teams, and more. To construct the QA dataset on structured EHR data, we conducted a poll at a university hospital and templatized the responses to create seed questions. Then, we manually linked them to two open-source EHR databases, MIMIC-III and eICU, and included them with various time expressions and held-out unanswerable questions in the dataset, which were all collected from the poll. Our dataset poses a unique set of challenges: the model needs to 1) generate SQL queries that reflect a wide range of needs in the hospital, including simple retrieval and complex operations such as calculating survival rate, 2) understand various time expressions to answer time-sensitive questions in healthcare, and 3) distinguish whether a given question is answerable or unanswerable based on the prediction confidence. We believe our dataset, EHRSQL, could serve as a practical benchmark to develop and assess QA models on structured EHR data and take one step further towards bridging the gap between text-to-SQL research and its real-life deployment in healthcare. EHRSQL is available at https://github.com/glee4810/EHRSQL.
Translation-equivariant Image Quantizer for Bi-directional Image-Text Generation
Dec 01, 2021



Abstract:Recently, vector-quantized image modeling has demonstrated impressive performance on generation tasks such as text-to-image generation. However, we discover that the current image quantizers do not satisfy translation equivariance in the quantized space due to aliasing, degrading performance in the downstream text-to-image generation and image-to-text generation, even in simple experimental setups. Instead of focusing on anti-aliasing, we take a direct approach to encourage translation equivariance in the quantized space. In particular, we explore a desirable property of image quantizers, called 'Translation Equivariance in the Quantized Space' and propose a simple but effective way to achieve translation equivariance by regularizing orthogonality in the codebook embedding vectors. Using this method, we improve accuracy by +22% in text-to-image generation and +26% in image-to-text generation, outperforming the VQGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge