Jiho Kim
Callie
SoK: DARPA's AI Cyber Challenge (AIxCC): Competition Design, Architectures, and Lessons Learned
Feb 07, 2026Abstract:DARPA's AI Cyber Challenge (AIxCC, 2023--2025) is the largest competition to date for building fully autonomous cyber reasoning systems (CRSs) that leverage recent advances in AI -- particularly large language models (LLMs) -- to discover and remediate vulnerabilities in real-world open-source software. This paper presents the first systematic analysis of AIxCC. Drawing on design documents, source code, execution traces, and discussions with organizers and competing teams, we examine the competition's structure and key design decisions, characterize the architectural approaches of finalist CRSs, and analyze competition results beyond the final scoreboard. Our analysis reveals the factors that truly drove CRS performance, identifies genuine technical advances achieved by teams, and exposes limitations that remain open for future research. We conclude with lessons for organizing future competitions and broader insights toward deploying autonomous CRSs in practice.
ECG-Agent: On-Device Tool-Calling Agent for ECG Multi-Turn Dialogue
Jan 28, 2026Abstract:Recent advances in Multimodal Large Language Models have rapidly expanded to electrocardiograms, focusing on classification, report generation, and single-turn QA tasks. However, these models fall short in real-world scenarios, lacking multi-turn conversational ability, on-device efficiency, and precise understanding of ECG measurements such as the PQRST intervals. To address these limitations, we introduce ECG-Agent, the first LLM-based tool-calling agent for multi-turn ECG dialogue. To facilitate its development and evaluation, we also present ECG-Multi-Turn-Dialogue (ECG-MTD) dataset, a collection of realistic user-assistant multi-turn dialogues for diverse ECG lead configurations. We develop ECG-Agents in various sizes, from on-device capable to larger agents. Experimental results show that ECG-Agents outperform baseline ECG-LLMs in response accuracy. Furthermore, on-device agents achieve comparable performance to larger agents in various evaluations that assess response accuracy, tool-calling ability, and hallucinations, demonstrating their viability for real-world applications.
ATLANTIS: AI-driven Threat Localization, Analysis, and Triage Intelligence System
Sep 18, 2025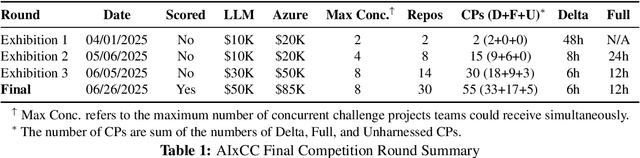
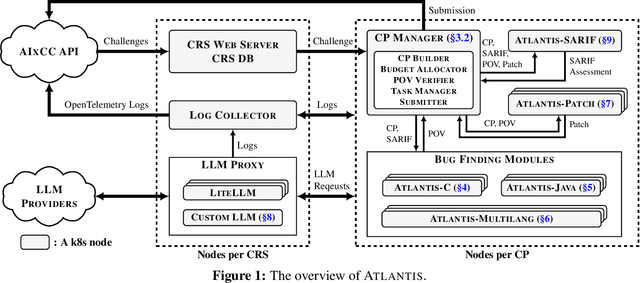
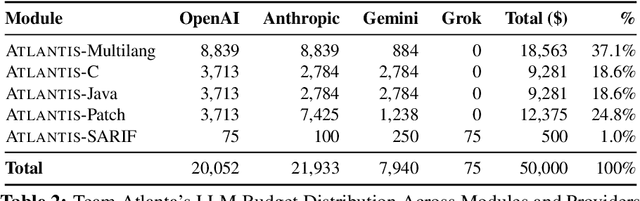
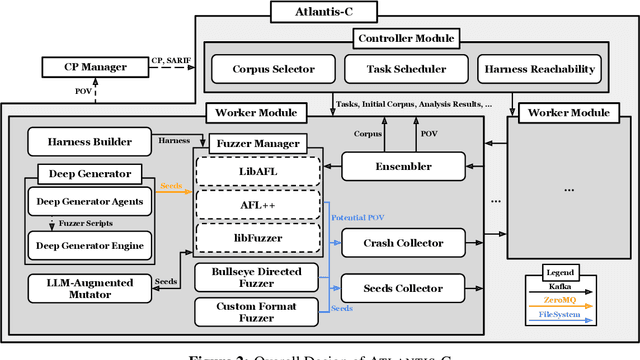
Abstract:We present ATLANTIS, the cyber reasoning system developed by Team Atlanta that won 1st place in the Final Competition of DARPA's AI Cyber Challenge (AIxCC) at DEF CON 33 (August 2025). AIxCC (2023-2025) challenged teams to build autonomous cyber reasoning systems capable of discovering and patching vulnerabilities at the speed and scale of modern software. ATLANTIS integrates large language models (LLMs) with program analysis -- combining symbolic execution, directed fuzzing, and static analysis -- to address limitations in automated vulnerability discovery and program repair. Developed by researchers at Georgia Institute of Technology, Samsung Research, KAIST, and POSTECH, the system addresses core challenges: scaling across diverse codebases from C to Java, achieving high precision while maintaining broad coverage, and producing semantically correct patches that preserve intended behavior. We detail the design philosophy, architectural decisions, and implementation strategies behind ATLANTIS, share lessons learned from pushing the boundaries of automated security when program analysis meets modern AI, and release artifacts to support reproducibility and future research.
PatientSim: A Persona-Driven Simulator for Realistic Doctor-Patient Interactions
May 23, 2025
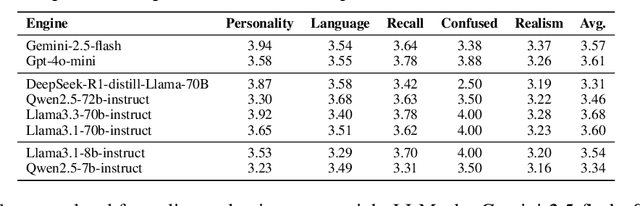
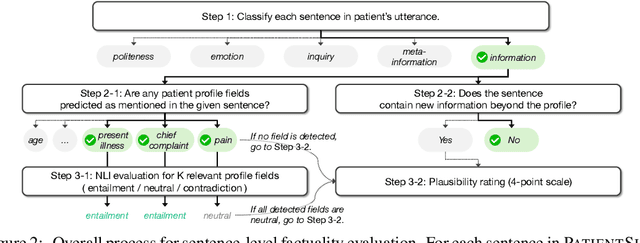
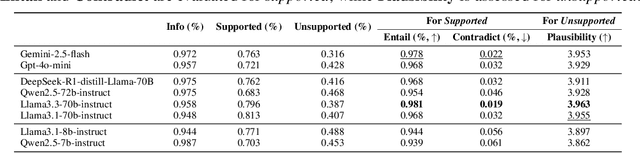
Abstract:Doctor-patient consultations require multi-turn, context-aware communication tailored to diverse patient personas. Training or evaluating doctor LLMs in such settings requires realistic patient interaction systems. However, existing simulators often fail to reflect the full range of personas seen in clinical practice. To address this, we introduce PatientSim, a patient simulator that generates realistic and diverse patient personas for clinical scenarios, grounded in medical expertise. PatientSim operates using: 1) clinical profiles, including symptoms and medical history, derived from real-world data in the MIMIC-ED and MIMIC-IV datasets, and 2) personas defined by four axes: personality, language proficiency, medical history recall level, and cognitive confusion level, resulting in 37 unique combinations. We evaluated eight LLMs for factual accuracy and persona consistency. The top-performing open-source model, Llama 3.3, was validated by four clinicians to confirm the robustness of our framework. As an open-source, customizable platform, PatientSim provides a reproducible and scalable solution that can be customized for specific training needs. Offering a privacy-compliant environment, it serves as a robust testbed for evaluating medical dialogue systems across diverse patient presentations and shows promise as an educational tool for healthcare.
Voice Interaction With Conversational AI Could Facilitate Thoughtful Reflection and Substantive Revision in Writing
Apr 11, 2025Abstract:Writing well requires not only expressing ideas but also refining them through revision, a process facilitated by reflection. Prior research suggests that feedback delivered through dialogues, such as those in writing center tutoring sessions, can help writers reflect more thoughtfully on their work compared to static feedback. Recent advancements in multi-modal large language models (LLMs) now offer new possibilities for supporting interactive and expressive voice-based reflection in writing. In particular, we propose that LLM-generated static feedback can be repurposed as conversation starters, allowing writers to seek clarification, request examples, and ask follow-up questions, thereby fostering deeper reflection on their writing. We argue that voice-based interaction can naturally facilitate this conversational exchange, encouraging writers' engagement with higher-order concerns, facilitating iterative refinement of their reflections, and reduce cognitive load compared to text-based interactions. To investigate these effects, we propose a formative study exploring how text vs. voice input influence writers' reflection and subsequent revisions. Findings from this study will inform the design of intelligent and interactive writing tools, offering insights into how voice-based interactions with LLM-powered conversational agents can support reflection and revision.
Design of a low-cost and lightweight 6 DoF bimanual arm for dynamic and contact-rich manipulation
Feb 24, 2025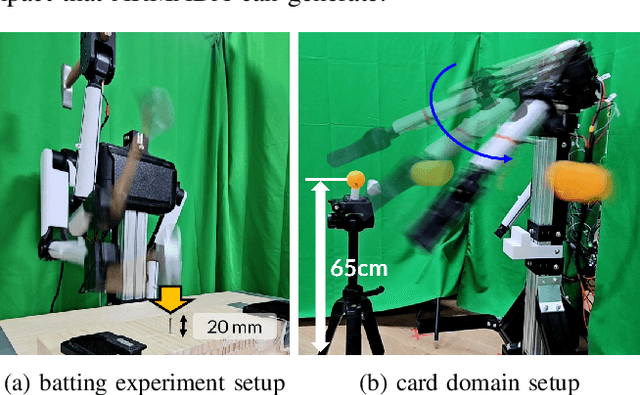
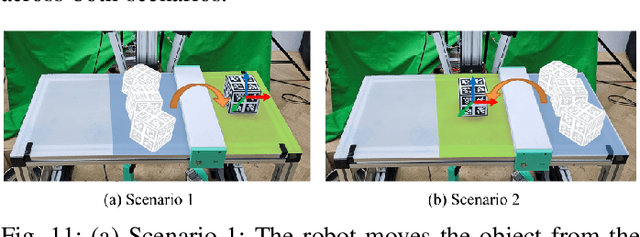
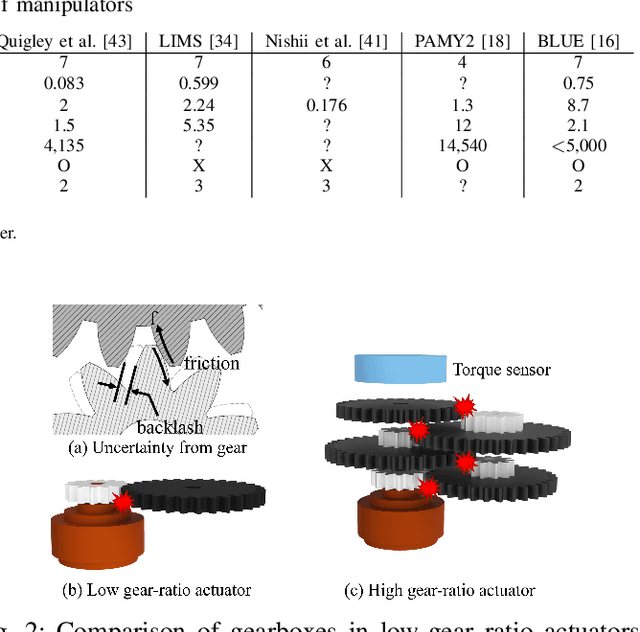
Abstract:Dynamic and contact-rich object manipulation, such as striking, snatching, or hammering, remains challenging for robotic systems due to hardware limitations. Most existing robots are constrained by high-inertia design, limited compliance, and reliance on expensive torque sensors. To address this, we introduce ARMADA (Affordable Robot for Manipulation and Dynamic Actions), a 6 degrees-of-freedom bimanual robot designed for dynamic manipulation research. ARMADA combines low-inertia, back-drivable actuators with a lightweight design, using readily available components and 3D-printed links for ease of assembly in research labs. The entire system, including both arms, is built for just $6,100. Each arm achieves speeds up to 6.16m/s, almost twice that of most collaborative robots, with a comparable payload of 2.5kg. We demonstrate ARMADA can perform dynamic manipulation like snatching, hammering, and bimanual throwing in real-world environments. We also showcase its effectiveness in reinforcement learning (RL) by training a non-prehensile manipulation policy in simulation and transferring it zero-shot to the real world, as well as human motion shadowing for dynamic bimanual object throwing. ARMADA is fully open-sourced with detailed assembly instructions, CAD models, URDFs, simulation, and learning codes. We highly recommend viewing the supplementary video at https://sites.google.com/view/im2-humanoid-arm.
R2-KG: General-Purpose Dual-Agent Framework for Reliable Reasoning on Knowledge Graphs
Feb 18, 2025



Abstract:Recent studies have combined Large Language Models (LLMs) with Knowledge Graphs (KGs) to enhance reasoning, improving inference accuracy without additional training while mitigating hallucination. However, existing frameworks are often rigid, struggling to adapt to KG or task changes. They also rely heavily on powerful LLMs for reliable (i.e., trustworthy) reasoning. To address this, We introduce R2-KG, a plug-and-play, dual-agent framework that separates reasoning into two roles: an Operator (a low-capacity LLM) that gathers evidence and a Supervisor (a high-capacity LLM) that makes final judgments. This design is cost-efficient for LLM inference while still maintaining strong reasoning accuracy. Additionally, R2-KG employs an Abstention mechanism, generating answers only when sufficient evidence is collected from KG, which significantly enhances reliability. Experiments across multiple KG-based reasoning tasks show that R2-KG consistently outperforms baselines in both accuracy and reliability, regardless of the inherent capability of LLMs used as the Operator. Further experiments reveal that the single-agent version of R2-KG, equipped with a strict self-consistency strategy, achieves significantly higher-than-baseline reliability while reducing inference cost. However, it also leads to a higher abstention rate in complex KGs. Our findings establish R2-KG as a flexible and cost-effective solution for KG-based reasoning. It reduces reliance on high-capacity LLMs while ensuring trustworthy inference.
Understanding the Performance and Estimating the Cost of LLM Fine-Tuning
Aug 08, 2024



Abstract:Due to the cost-prohibitive nature of training Large Language Models (LLMs), fine-tuning has emerged as an attractive alternative for specializing LLMs for specific tasks using limited compute resources in a cost-effective manner. In this paper, we characterize sparse Mixture of Experts (MoE) based LLM fine-tuning to understand their accuracy and runtime performance on a single GPU. Our evaluation provides unique insights into the training efficacy of sparse and dense versions of MoE models, as well as their runtime characteristics, including maximum batch size, execution time breakdown, end-to-end throughput, GPU hardware utilization, and load distribution. Our study identifies the optimization of the MoE layer as crucial for further improving the performance of LLM fine-tuning. Using our profiling results, we also develop and validate an analytical model to estimate the cost of LLM fine-tuning on the cloud. This model, based on parameters of the model and GPU architecture, estimates LLM throughput and the cost of training, aiding practitioners in industry and academia to budget the cost of fine-tuning a specific model.
EHRCon: Dataset for Checking Consistency between Unstructured Notes and Structured Tables in Electronic Health Records
Jun 24, 2024



Abstract:Electronic Health Records (EHRs) are integral for storing comprehensive patient medical records, combining structured data (e.g., medications) with detailed clinical notes (e.g., physician notes). These elements are essential for straightforward data retrieval and provide deep, contextual insights into patient care. However, they often suffer from discrepancies due to unintuitive EHR system designs and human errors, posing serious risks to patient safety. To address this, we developed EHRCon, a new dataset and task specifically designed to ensure data consistency between structured tables and unstructured notes in EHRs. EHRCon was crafted in collaboration with healthcare professionals using the MIMIC-III EHR dataset, and includes manual annotations of 3,943 entities across 105 clinical notes checked against database entries for consistency. EHRCon has two versions, one using the original MIMIC-III schema, and another using the OMOP CDM schema, in order to increase its applicability and generalizability. Furthermore, leveraging the capabilities of large language models, we introduce CheckEHR, a novel framework for verifying the consistency between clinical notes and database tables. CheckEHR utilizes an eight-stage process and shows promising results in both few-shot and zero-shot settings. The code is available at https://github.com/dustn1259/EHRCon.
DialSim: A Real-Time Simulator for Evaluating Long-Term Dialogue Understanding of Conversational Agents
Jun 19, 2024



Abstract:Recent advancements in Large Language Models (LLMs) have significantly enhanced the capabilities of conversational agents, making them applicable to various fields (e.g., education). Despite their progress, the evaluation of the agents often overlooks the complexities of real-world conversations, such as real-time interactions, multi-party dialogues, and extended contextual dependencies. To bridge this gap, we introduce DialSim, a real-time dialogue simulator. In this simulator, an agent is assigned the role of a character from popular TV shows, requiring it to respond to spontaneous questions using past dialogue information and to distinguish between known and unknown information. Key features of DialSim include evaluating the agent's ability to respond within a reasonable time limit, handling long-term multi-party dialogues, and managing adversarial settings (e.g., swap character names) to challenge the agent's reliance on pre-trained knowledge. We utilized this simulator to evaluate the latest conversational agents and analyze their limitations. Our experiments highlight both the strengths and weaknesses of these agents, providing valuable insights for future improvements in the field of conversational AI. DialSim is available at https://github.com/jiho283/Simulator.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge