Kihyun You
CheX-GPT: Harnessing Large Language Models for Enhanced Chest X-ray Report Labeling
Jan 21, 2024



Abstract:Free-text radiology reports present a rich data source for various medical tasks, but effectively labeling these texts remains challenging. Traditional rule-based labeling methods fall short of capturing the nuances of diverse free-text patterns. Moreover, models using expert-annotated data are limited by data scarcity and pre-defined classes, impacting their performance, flexibility and scalability. To address these issues, our study offers three main contributions: 1) We demonstrate the potential of GPT as an adept labeler using carefully designed prompts. 2) Utilizing only the data labeled by GPT, we trained a BERT-based labeler, CheX-GPT, which operates faster and more efficiently than its GPT counterpart. 3) To benchmark labeler performance, we introduced a publicly available expert-annotated test set, MIMIC-500, comprising 500 cases from the MIMIC validation set. Our findings demonstrate that CheX-GPT not only excels in labeling accuracy over existing models, but also showcases superior efficiency, flexibility, and scalability, supported by our introduction of the MIMIC-500 dataset for robust benchmarking. Code and models are available at https://github.com/kakaobrain/CheXGPT.
CXR-CLIP: Toward Large Scale Chest X-ray Language-Image Pre-training
Oct 20, 2023Abstract:A large-scale image-text pair dataset has greatly contributed to the development of vision-language pre-training (VLP) models, which enable zero-shot or few-shot classification without costly annotation. However, in the medical domain, the scarcity of data remains a significant challenge for developing a powerful VLP model. In this paper, we tackle the lack of image-text data in chest X-ray by expanding image-label pair as image-text pair via general prompt and utilizing multiple images and multiple sections in a radiologic report. We also design two contrastive losses, named ICL and TCL, for learning study-level characteristics of medical images and reports, respectively. Our model outperforms the state-of-the-art models trained under the same conditions. Also, enlarged dataset improve the discriminative power of our pre-trained model for classification, while sacrificing marginal retrieval performance. Code is available at https://github.com/kakaobrain/cxr-clip.
Diversified Mutual Learning for Deep Metric Learning
Sep 09, 2020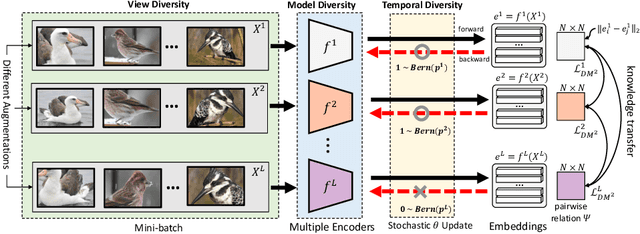
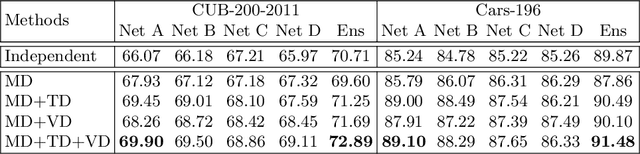
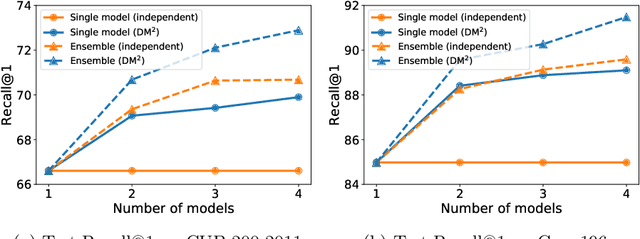
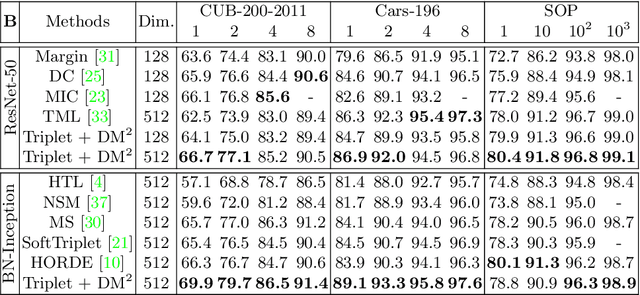
Abstract:Mutual learning is an ensemble training strategy to improve generalization by transferring individual knowledge to each other while simultaneously training multiple models. In this work, we propose an effective mutual learning method for deep metric learning, called Diversified Mutual Metric Learning, which enhances embedding models with diversified mutual learning. We transfer relational knowledge for deep metric learning by leveraging three kinds of diversities in mutual learning: (1) model diversity from different initializations of models, (2) temporal diversity from different frequencies of parameter update, and (3) view diversity from different augmentations of inputs. Our method is particularly adequate for inductive transfer learning at the lack of large-scale data, where the embedding model is initialized with a pretrained model and then fine-tuned on a target dataset. Extensive experiments show that our method significantly improves individual models as well as their ensemble. Finally, the proposed method with a conventional triplet loss achieves the state-of-the-art performance of Recall@1 on standard datasets: 69.9 on CUB-200-2011 and 89.1 on CARS-196.
Real-Time Object Tracking via Meta-Learning: Efficient Model Adaptation and One-Shot Channel Pruning
Dec 04, 2019



Abstract:We propose a novel meta-learning framework for real-time object tracking with efficient model adaptation and channel pruning. Given an object tracker, our framework learns to fine-tune its model parameters in only a few iterations of gradient-descent during tracking while pruning its network channels using the target ground-truth at the first frame. Such a learning problem is formulated as a meta-learning task, where a meta-tracker is trained by updating its meta-parameters for initial weights, learning rates, and pruning masks through carefully designed tracking simulations. The integrated meta-tracker greatly improves tracking performance by accelerating the convergence of online learning and reducing the cost of feature computation. Experimental evaluation on the standard datasets demonstrates its outstanding accuracy and speed compared to the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge