Beomhee Park
CXR-CLIP: Toward Large Scale Chest X-ray Language-Image Pre-training
Oct 20, 2023Abstract:A large-scale image-text pair dataset has greatly contributed to the development of vision-language pre-training (VLP) models, which enable zero-shot or few-shot classification without costly annotation. However, in the medical domain, the scarcity of data remains a significant challenge for developing a powerful VLP model. In this paper, we tackle the lack of image-text data in chest X-ray by expanding image-label pair as image-text pair via general prompt and utilizing multiple images and multiple sections in a radiologic report. We also design two contrastive losses, named ICL and TCL, for learning study-level characteristics of medical images and reports, respectively. Our model outperforms the state-of-the-art models trained under the same conditions. Also, enlarged dataset improve the discriminative power of our pre-trained model for classification, while sacrificing marginal retrieval performance. Code is available at https://github.com/kakaobrain/cxr-clip.
The Medical Segmentation Decathlon
Jun 10, 2021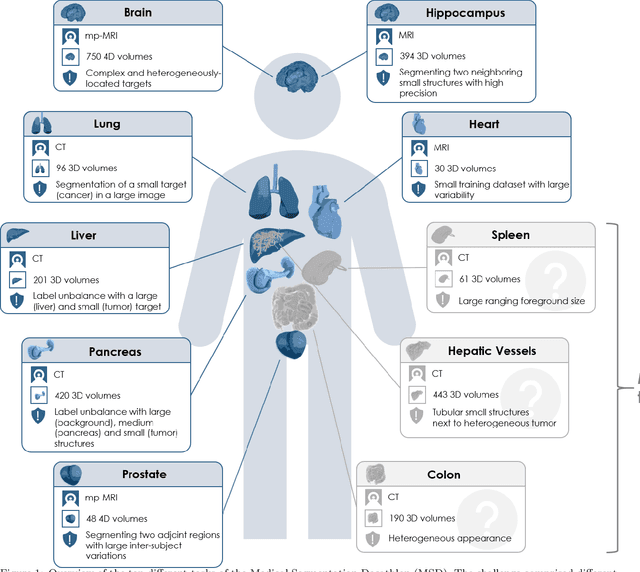
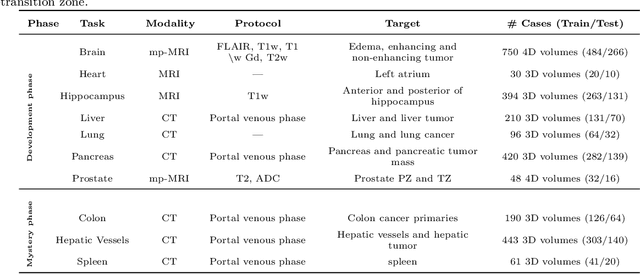
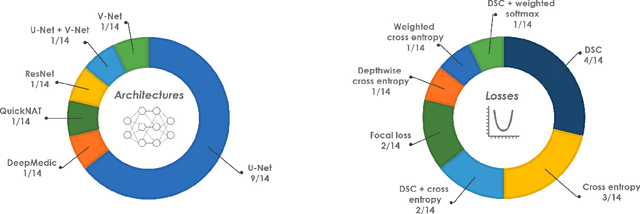
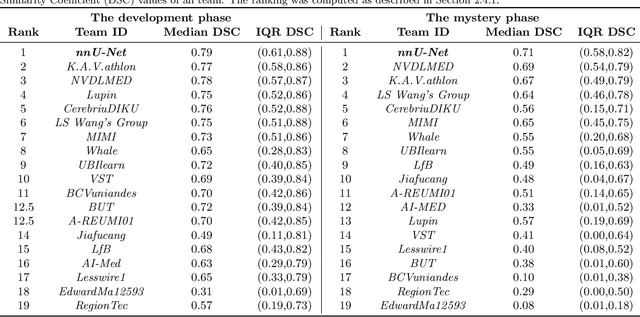
Abstract:International challenges have become the de facto standard for comparative assessment of image analysis algorithms given a specific task. Segmentation is so far the most widely investigated medical image processing task, but the various segmentation challenges have typically been organized in isolation, such that algorithm development was driven by the need to tackle a single specific clinical problem. We hypothesized that a method capable of performing well on multiple tasks will generalize well to a previously unseen task and potentially outperform a custom-designed solution. To investigate the hypothesis, we organized the Medical Segmentation Decathlon (MSD) - a biomedical image analysis challenge, in which algorithms compete in a multitude of both tasks and modalities. The underlying data set was designed to explore the axis of difficulties typically encountered when dealing with medical images, such as small data sets, unbalanced labels, multi-site data and small objects. The MSD challenge confirmed that algorithms with a consistent good performance on a set of tasks preserved their good average performance on a different set of previously unseen tasks. Moreover, by monitoring the MSD winner for two years, we found that this algorithm continued generalizing well to a wide range of other clinical problems, further confirming our hypothesis. Three main conclusions can be drawn from this study: (1) state-of-the-art image segmentation algorithms are mature, accurate, and generalize well when retrained on unseen tasks; (2) consistent algorithmic performance across multiple tasks is a strong surrogate of algorithmic generalizability; (3) the training of accurate AI segmentation models is now commoditized to non AI experts.
Resource Optimized Neural Architecture Search for 3D Medical Image Segmentation
Sep 02, 2019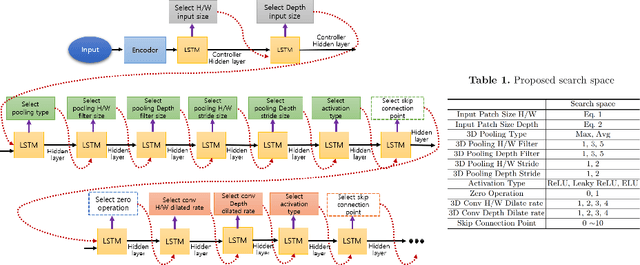
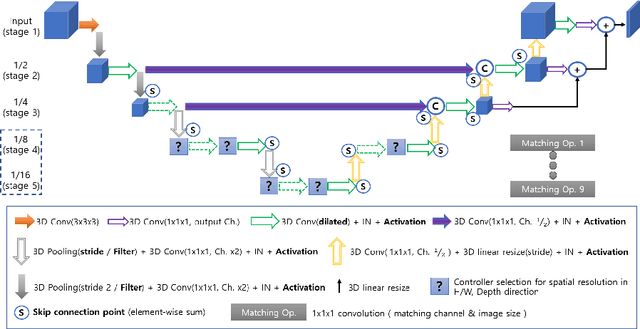
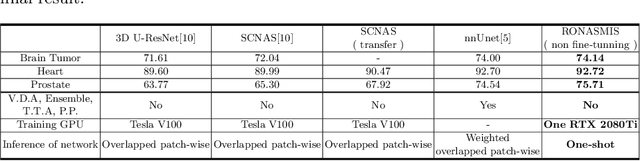
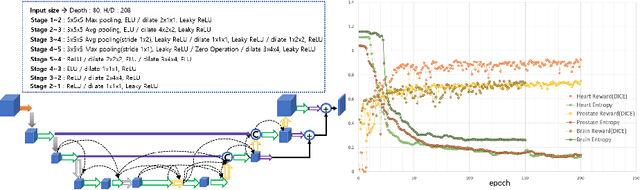
Abstract:Neural Architecture Search (NAS), a framework which automates the task of designing neural networks, has recently been actively studied in the field of deep learning. However, there are only a few NAS methods suitable for 3D medical image segmentation. Medical 3D images are generally very large; thus it is difficult to apply previous NAS methods due to their GPU computational burden and long training time. We propose the resource-optimized neural architecture search method which can be applied to 3D medical segmentation tasks in a short training time (1.39 days for 1GB dataset) using a small amount of computation power (one RTX 2080Ti, 10.8GB GPU memory). Excellent performance can also be achieved without retraining(fine-tuning) which is essential in most NAS methods. These advantages can be achieved by using a reinforcement learning-based controller with parameter sharing and focusing on the optimal search space configuration of macro search rather than micro search. Our experiments demonstrate that the proposed NAS method outperforms manually designed networks with state-of-the-art performance in 3D medical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge