Yeonsu Kwon
ECG-Agent: On-Device Tool-Calling Agent for ECG Multi-Turn Dialogue
Jan 28, 2026Abstract:Recent advances in Multimodal Large Language Models have rapidly expanded to electrocardiograms, focusing on classification, report generation, and single-turn QA tasks. However, these models fall short in real-world scenarios, lacking multi-turn conversational ability, on-device efficiency, and precise understanding of ECG measurements such as the PQRST intervals. To address these limitations, we introduce ECG-Agent, the first LLM-based tool-calling agent for multi-turn ECG dialogue. To facilitate its development and evaluation, we also present ECG-Multi-Turn-Dialogue (ECG-MTD) dataset, a collection of realistic user-assistant multi-turn dialogues for diverse ECG lead configurations. We develop ECG-Agents in various sizes, from on-device capable to larger agents. Experimental results show that ECG-Agents outperform baseline ECG-LLMs in response accuracy. Furthermore, on-device agents achieve comparable performance to larger agents in various evaluations that assess response accuracy, tool-calling ability, and hallucinations, demonstrating their viability for real-world applications.
Time is Not Enough: Time-Frequency based Explanation for Time-Series Black-Box Models
Aug 07, 2024



Abstract:Despite the massive attention given to time-series explanations due to their extensive applications, a notable limitation in existing approaches is their primary reliance on the time-domain. This overlooks the inherent characteristic of time-series data containing both time and frequency features. In this work, we present Spectral eXplanation (SpectralX), an XAI framework that provides time-frequency explanations for time-series black-box classifiers. This easily adaptable framework enables users to "plug-in" various perturbation-based XAI methods for any pre-trained time-series classification models to assess their impact on the explanation quality without having to modify the framework architecture. Additionally, we introduce Feature Importance Approximations (FIA), a new perturbation-based XAI method. These methods consist of feature insertion, deletion, and combination techniques to enhance computational efficiency and class-specific explanations in time-series classification tasks. We conduct extensive experiments in the generated synthetic dataset and various UCR Time-Series datasets to first compare the explanation performance of FIA and other existing perturbation-based XAI methods in both time-domain and time-frequency domain, and then show the superiority of our FIA in the time-frequency domain with the SpectralX framework. Finally, we conduct a user study to confirm the practicality of our FIA in SpectralX framework for class-specific time-frequency based time-series explanations. The source code is available in https://github.com/gustmd0121/Time_is_not_Enough
EHRCon: Dataset for Checking Consistency between Unstructured Notes and Structured Tables in Electronic Health Records
Jun 24, 2024



Abstract:Electronic Health Records (EHRs) are integral for storing comprehensive patient medical records, combining structured data (e.g., medications) with detailed clinical notes (e.g., physician notes). These elements are essential for straightforward data retrieval and provide deep, contextual insights into patient care. However, they often suffer from discrepancies due to unintuitive EHR system designs and human errors, posing serious risks to patient safety. To address this, we developed EHRCon, a new dataset and task specifically designed to ensure data consistency between structured tables and unstructured notes in EHRs. EHRCon was crafted in collaboration with healthcare professionals using the MIMIC-III EHR dataset, and includes manual annotations of 3,943 entities across 105 clinical notes checked against database entries for consistency. EHRCon has two versions, one using the original MIMIC-III schema, and another using the OMOP CDM schema, in order to increase its applicability and generalizability. Furthermore, leveraging the capabilities of large language models, we introduce CheckEHR, a novel framework for verifying the consistency between clinical notes and database tables. CheckEHR utilizes an eight-stage process and shows promising results in both few-shot and zero-shot settings. The code is available at https://github.com/dustn1259/EHRCon.
KG-GPT: A General Framework for Reasoning on Knowledge Graphs Using Large Language Models
Oct 17, 2023



Abstract:While large language models (LLMs) have made considerable advancements in understanding and generating unstructured text, their application in structured data remains underexplored. Particularly, using LLMs for complex reasoning tasks on knowledge graphs (KGs) remains largely untouched. To address this, we propose KG-GPT, a multi-purpose framework leveraging LLMs for tasks employing KGs. KG-GPT comprises three steps: Sentence Segmentation, Graph Retrieval, and Inference, each aimed at partitioning sentences, retrieving relevant graph components, and deriving logical conclusions, respectively. We evaluate KG-GPT using KG-based fact verification and KGQA benchmarks, with the model showing competitive and robust performance, even outperforming several fully-supervised models. Our work, therefore, marks a significant step in unifying structured and unstructured data processing within the realm of LLMs.
FactKG: Fact Verification via Reasoning on Knowledge Graphs
May 19, 2023Abstract:In real world applications, knowledge graphs (KG) are widely used in various domains (e.g. medical applications and dialogue agents). However, for fact verification, KGs have not been adequately utilized as a knowledge source. KGs can be a valuable knowledge source in fact verification due to their reliability and broad applicability. A KG consists of nodes and edges which makes it clear how concepts are linked together, allowing machines to reason over chains of topics. However, there are many challenges in understanding how these machine-readable concepts map to information in text. To enable the community to better use KGs, we introduce a new dataset, FactKG: Fact Verification via Reasoning on Knowledge Graphs. It consists of 108k natural language claims with five types of reasoning: One-hop, Conjunction, Existence, Multi-hop, and Negation. Furthermore, FactKG contains various linguistic patterns, including colloquial style claims as well as written style claims to increase practicality. Lastly, we develop a baseline approach and analyze FactKG over these reasoning types. We believe FactKG can advance both reliability and practicality in KG-based fact verification.
Open-WikiTable: Dataset for Open Domain Question Answering with Complex Reasoning over Table
May 12, 2023
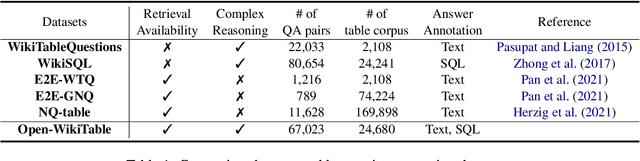


Abstract:Despite recent interest in open domain question answering (ODQA) over tables, many studies still rely on datasets that are not truly optimal for the task with respect to utilizing structural nature of table. These datasets assume answers reside as a single cell value and do not necessitate exploring over multiple cells such as aggregation, comparison, and sorting. Thus, we release Open-WikiTable, the first ODQA dataset that requires complex reasoning over tables. Open-WikiTable is built upon WikiSQL and WikiTableQuestions to be applicable in the open-domain setting. As each question is coupled with both textual answers and SQL queries, Open-WikiTable opens up a wide range of possibilities for future research, as both reader and parser methods can be applied. The dataset and code are publicly available.
EHRSQL: A Practical Text-to-SQL Benchmark for Electronic Health Records
Jan 16, 2023



Abstract:We present a new text-to-SQL dataset for electronic health records (EHRs). The utterances were collected from 222 hospital staff, including physicians, nurses, insurance review and health records teams, and more. To construct the QA dataset on structured EHR data, we conducted a poll at a university hospital and templatized the responses to create seed questions. Then, we manually linked them to two open-source EHR databases, MIMIC-III and eICU, and included them with various time expressions and held-out unanswerable questions in the dataset, which were all collected from the poll. Our dataset poses a unique set of challenges: the model needs to 1) generate SQL queries that reflect a wide range of needs in the hospital, including simple retrieval and complex operations such as calculating survival rate, 2) understand various time expressions to answer time-sensitive questions in healthcare, and 3) distinguish whether a given question is answerable or unanswerable based on the prediction confidence. We believe our dataset, EHRSQL, could serve as a practical benchmark to develop and assess QA models on structured EHR data and take one step further towards bridging the gap between text-to-SQL research and its real-life deployment in healthcare. EHRSQL is available at https://github.com/glee4810/EHRSQL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge