Tackeun Kim
Towards Predicting Temporal Changes in a Patient's Chest X-ray Images based on Electronic Health Records
Sep 11, 2024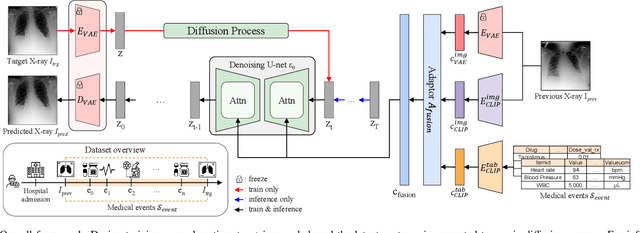
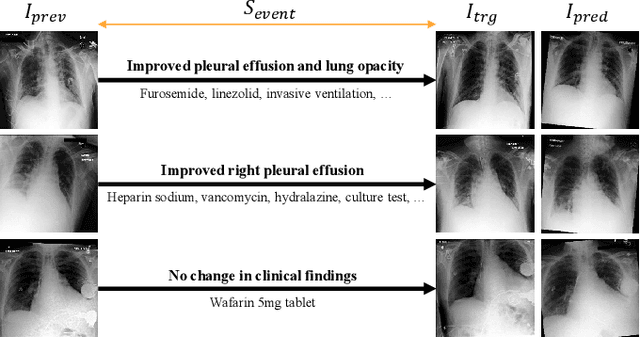
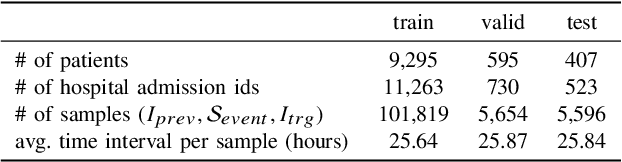
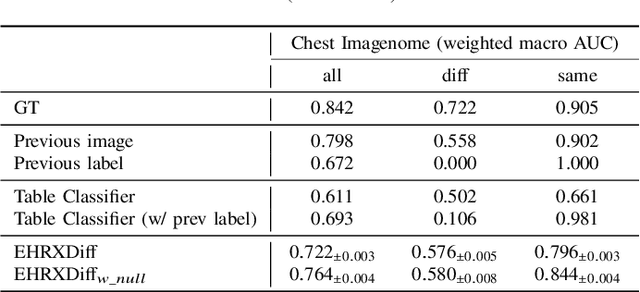
Abstract:Chest X-ray imaging (CXR) is an important diagnostic tool used in hospitals to assess patient conditions and monitor changes over time. Generative models, specifically diffusion-based models, have shown promise in generating realistic synthetic X-rays. However, these models mainly focus on conditional generation using single-time-point data, i.e., typically CXRs taken at a specific time with their corresponding reports, limiting their clinical utility, particularly for capturing temporal changes. To address this limitation, we propose a novel framework, EHRXDiff, which predicts future CXR images by integrating previous CXRs with subsequent medical events, e.g., prescriptions, lab measures, etc. Our framework dynamically tracks and predicts disease progression based on a latent diffusion model, conditioned on the previous CXR image and a history of medical events. We comprehensively evaluate the performance of our framework across three key aspects, including clinical consistency, demographic consistency, and visual realism. We demonstrate that our framework generates high-quality, realistic future images that capture potential temporal changes, suggesting its potential for further development as a clinical simulation tool. This could offer valuable insights for patient monitoring and treatment planning in the medical field.
EHRXQA: A Multi-Modal Question Answering Dataset for Electronic Health Records with Chest X-ray Images
Oct 28, 2023



Abstract:Electronic Health Records (EHRs), which contain patients' medical histories in various multi-modal formats, often overlook the potential for joint reasoning across imaging and table modalities underexplored in current EHR Question Answering (QA) systems. In this paper, we introduce EHRXQA, a novel multi-modal question answering dataset combining structured EHRs and chest X-ray images. To develop our dataset, we first construct two uni-modal resources: 1) The MIMIC- CXR-VQA dataset, our newly created medical visual question answering (VQA) benchmark, specifically designed to augment the imaging modality in EHR QA, and 2) EHRSQL (MIMIC-IV), a refashioned version of a previously established table-based EHR QA dataset. By integrating these two uni-modal resources, we successfully construct a multi-modal EHR QA dataset that necessitates both uni-modal and cross-modal reasoning. To address the unique challenges of multi-modal questions within EHRs, we propose a NeuralSQL-based strategy equipped with an external VQA API. This pioneering endeavor enhances engagement with multi-modal EHR sources and we believe that our dataset can catalyze advances in real-world medical scenarios such as clinical decision-making and research. EHRXQA is available at https://github.com/baeseongsu/ehrxqa.
Unified Chest X-ray and Radiology Report Generation Model with Multi-view Chest X-rays
Mar 01, 2023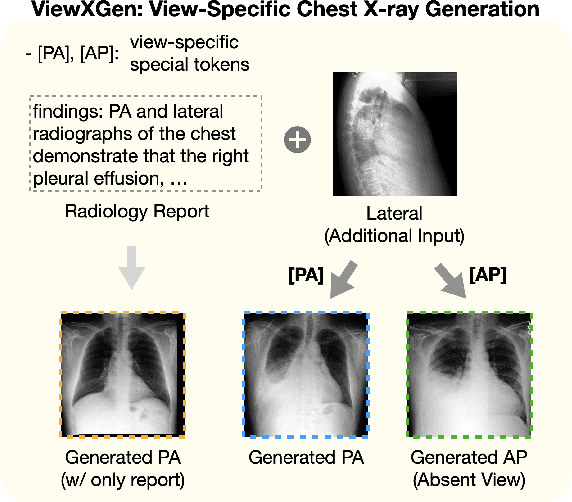
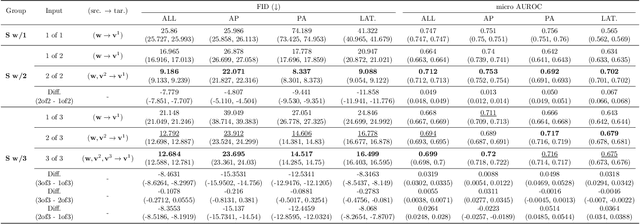
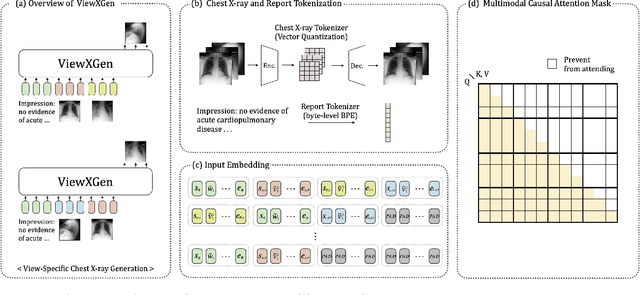
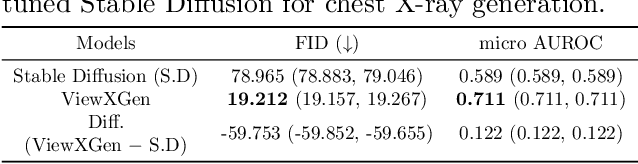
Abstract:Generated synthetic data in medical research can substitute privacy and security-sensitive data with a large-scale curated dataset, reducing data collection and annotation costs. As part of this effort, we propose UniXGen, a unified chest X-ray and report generation model, with the following contributions. First, we design a unified model for bidirectional chest X-ray and report generation by adopting a vector quantization method to discretize chest X-rays into discrete visual tokens and formulating both tasks as sequence generation tasks. Second, we introduce several special tokens to generate chest X-rays with specific views that can be useful when the desired views are unavailable. Furthermore, UniXGen can flexibly take various inputs from single to multiple views to take advantage of the additional findings available in other X-ray views. We adopt an efficient transformer for computational and memory efficiency to handle the long-range input sequence of multi-view chest X-rays with high resolution and long paragraph reports. In extensive experiments, we show that our unified model has a synergistic effect on both generation tasks, as opposed to training only the task-specific models. We also find that view-specific special tokens can distinguish between different views and properly generate specific views even if they do not exist in the dataset, and utilizing multi-view chest X-rays can faithfully capture the abnormal findings in the additional X-rays. The source code is publicly available at: https://github.com/ttumyche/UniXGen.
Graph-Text Multi-Modal Pre-training for Medical Representation Learning
Mar 18, 2022



Abstract:As the volume of Electronic Health Records (EHR) sharply grows, there has been emerging interest in learning the representation of EHR for healthcare applications. Representation learning of EHR requires appropriate modeling of the two dominant modalities in EHR: structured data and unstructured text. In this paper, we present MedGTX, a pre-trained model for multi-modal representation learning of the structured and textual EHR data. MedGTX uses a novel graph encoder to exploit the graphical nature of structured EHR data, and a text encoder to handle unstructured text, and a cross-modal encoder to learn a joint representation space. We pre-train our model through four proxy tasks on MIMIC-III, an open-source EHR data, and evaluate our model on two clinical benchmarks and three novel downstream tasks which tackle real-world problems in EHR data. The results consistently show the effectiveness of pre-training the model for joint representation of both structured and unstructured information from EHR. Given the promising performance of MedGTX, we believe this work opens a new door to jointly understanding the two fundamental modalities of EHR data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge