Jing Ke
Generating Event-oriented Attribution for Movies via Two-Stage Prefix-Enhanced Multimodal LLM
Sep 14, 2024Abstract:The prosperity of social media platforms has raised the urgent demand for semantic-rich services, e.g., event and storyline attribution. However, most existing research focuses on clip-level event understanding, primarily through basic captioning tasks, without analyzing the causes of events across an entire movie. This is a significant challenge, as even advanced multimodal large language models (MLLMs) struggle with extensive multimodal information due to limited context length. To address this issue, we propose a Two-Stage Prefix-Enhanced MLLM (TSPE) approach for event attribution, i.e., connecting associated events with their causal semantics, in movie videos. In the local stage, we introduce an interaction-aware prefix that guides the model to focus on the relevant multimodal information within a single clip, briefly summarizing the single event. Correspondingly, in the global stage, we strengthen the connections between associated events using an inferential knowledge graph, and design an event-aware prefix that directs the model to focus on associated events rather than all preceding clips, resulting in accurate event attribution. Comprehensive evaluations of two real-world datasets demonstrate that our framework outperforms state-of-the-art methods.
A Survey for Large Language Models in Biomedicine
Aug 29, 2024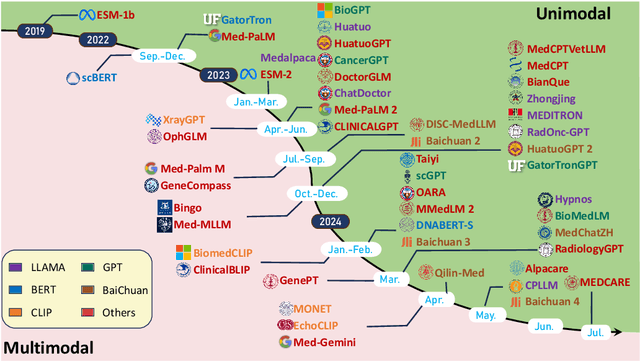
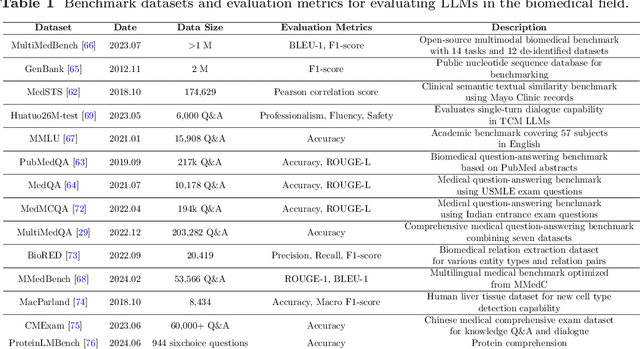
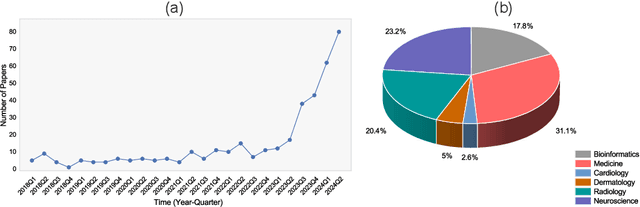
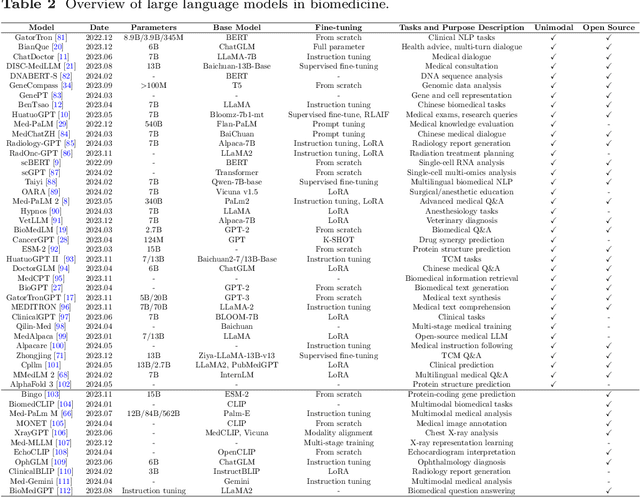
Abstract:Recent breakthroughs in large language models (LLMs) offer unprecedented natural language understanding and generation capabilities. However, existing surveys on LLMs in biomedicine often focus on specific applications or model architectures, lacking a comprehensive analysis that integrates the latest advancements across various biomedical domains. This review, based on an analysis of 484 publications sourced from databases including PubMed, Web of Science, and arXiv, provides an in-depth examination of the current landscape, applications, challenges, and prospects of LLMs in biomedicine, distinguishing itself by focusing on the practical implications of these models in real-world biomedical contexts. Firstly, we explore the capabilities of LLMs in zero-shot learning across a broad spectrum of biomedical tasks, including diagnostic assistance, drug discovery, and personalized medicine, among others, with insights drawn from 137 key studies. Then, we discuss adaptation strategies of LLMs, including fine-tuning methods for both uni-modal and multi-modal LLMs to enhance their performance in specialized biomedical contexts where zero-shot fails to achieve, such as medical question answering and efficient processing of biomedical literature. Finally, we discuss the challenges that LLMs face in the biomedicine domain including data privacy concerns, limited model interpretability, issues with dataset quality, and ethics due to the sensitive nature of biomedical data, the need for highly reliable model outputs, and the ethical implications of deploying AI in healthcare. To address these challenges, we also identify future research directions of LLM in biomedicine including federated learning methods to preserve data privacy and integrating explainable AI methodologies to enhance the transparency of LLMs.
DiffImpute: Tabular Data Imputation With Denoising Diffusion Probabilistic Model
Mar 20, 2024Abstract:Tabular data plays a crucial role in various domains but often suffers from missing values, thereby curtailing its potential utility. Traditional imputation techniques frequently yield suboptimal results and impose substantial computational burdens, leading to inaccuracies in subsequent modeling tasks. To address these challenges, we propose DiffImpute, a novel Denoising Diffusion Probabilistic Model (DDPM). Specifically, DiffImpute is trained on complete tabular datasets, ensuring that it can produce credible imputations for missing entries without undermining the authenticity of the existing data. Innovatively, it can be applied to various settings of Missing Completely At Random (MCAR) and Missing At Random (MAR). To effectively handle the tabular features in DDPM, we tailor four tabular denoising networks, spanning MLP, ResNet, Transformer, and U-Net. We also propose Harmonization to enhance coherence between observed and imputed data by infusing the data back and denoising them multiple times during the sampling stage. To enable efficient inference while maintaining imputation performance, we propose a refined non-Markovian sampling process that works along with Harmonization. Empirical evaluations on seven diverse datasets underscore the prowess of DiffImpute. Specifically, when paired with the Transformer as the denoising network, it consistently outperforms its competitors, boasting an average ranking of 1.7 and the most minimal standard deviation. In contrast, the next best method lags with a ranking of 2.8 and a standard deviation of 0.9. The code is available at https://github.com/Dendiiiii/DiffImpute.
TransNuSeg: A Lightweight Multi-Task Transformer for Nuclei Segmentation
Jul 16, 2023Abstract:Nuclei appear small in size, yet, in real clinical practice, the global spatial information and correlation of the color or brightness contrast between nuclei and background, have been considered a crucial component for accurate nuclei segmentation. However, the field of automatic nuclei segmentation is dominated by Convolutional Neural Networks (CNNs), meanwhile, the potential of the recently prevalent Transformers has not been fully explored, which is powerful in capturing local-global correlations. To this end, we make the first attempt at a pure Transformer framework for nuclei segmentation, called TransNuSeg. Different from prior work, we decouple the challenging nuclei segmentation task into an intrinsic multi-task learning task, where a tri-decoder structure is employed for nuclei instance, nuclei edge, and clustered edge segmentation respectively. To eliminate the divergent predictions from different branches in previous work, a novel self distillation loss is introduced to explicitly impose consistency regulation between branches. Moreover, to formulate the high correlation between branches and also reduce the number of parameters, an efficient attention sharing scheme is proposed by partially sharing the self-attention heads amongst the tri-decoders. Finally, a token MLP bottleneck replaces the over-parameterized Transformer bottleneck for a further reduction in model complexity. Experiments on two datasets of different modalities, including MoNuSeg have shown that our methods can outperform state-of-the-art counterparts such as CA2.5-Net by 2-3% Dice with 30% fewer parameters. In conclusion, TransNuSeg confirms the strength of Transformer in the context of nuclei segmentation, which thus can serve as an efficient solution for real clinical practice. Code is available at https://github.com/zhenqi-he/transnuseg.
RandStainNA: Learning Stain-Agnostic Features from Histology Slides by Bridging Stain Augmentation and Normalization
Jun 25, 2022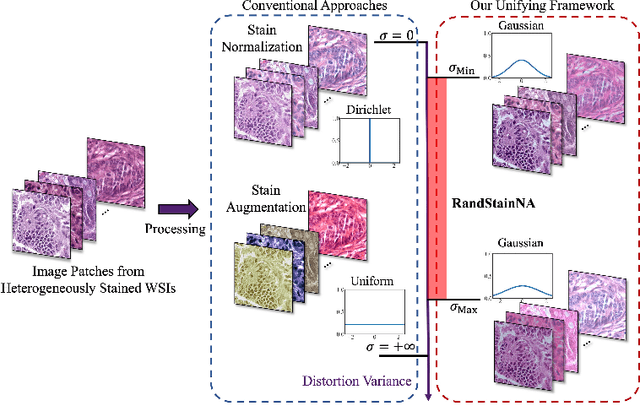
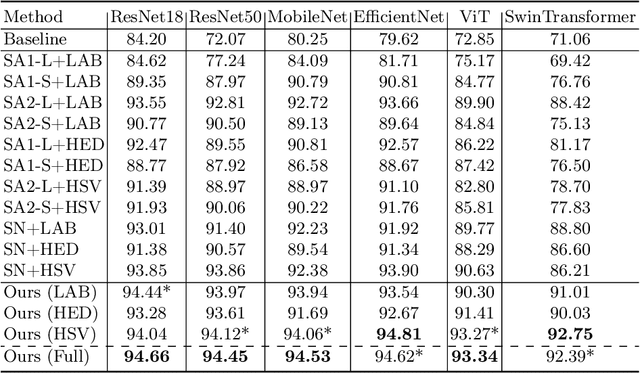
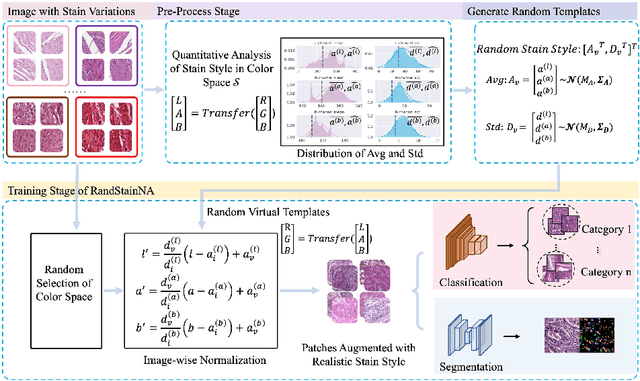

Abstract:Stain variations often decrease the generalization ability of deep learning based approaches in digital histopathology analysis. Two separate proposals, namely stain normalization (SN) and stain augmentation (SA), have been spotlighted to reduce the generalization error, where the former alleviates the stain shift across different medical centers using template image and the latter enriches the accessible stain styles by the simulation of more stain variations. However, their applications are bounded by the selection of template images and the construction of unrealistic styles. To address the problems, we unify SN and SA with a novel RandStainNA scheme, which constrains variable stain styles in a practicable range to train a stain agnostic deep learning model. The RandStainNA is applicable to stain normalization in a collection of color spaces i.e. HED, HSV, LAB. Additionally, we propose a random color space selection scheme to gain extra performance improvement. We evaluate our method by two diagnostic tasks i.e. tissue subtype classification and nuclei segmentation, with various network backbones. The performance superiority over both SA and SN yields that the proposed RandStainNA can consistently improve the generalization ability, that our models can cope with more incoming clinical datasets with unpredicted stain styles. The codes is available at https://github.com/yiqings/RandStainNA.
Approximate Random Dropout
May 23, 2018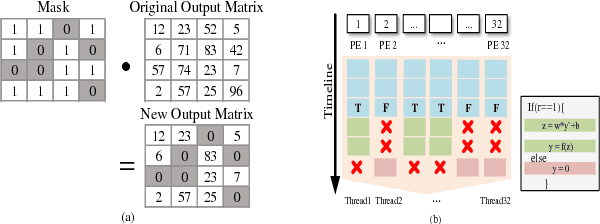
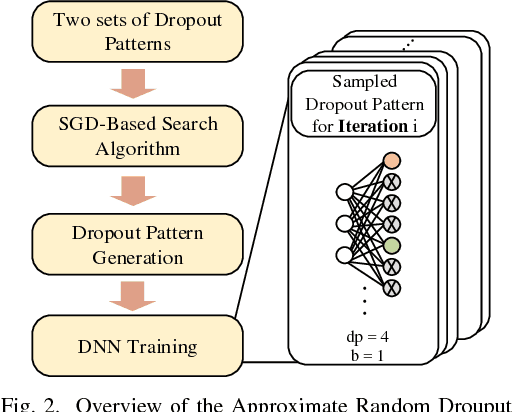
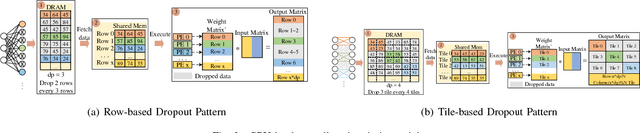
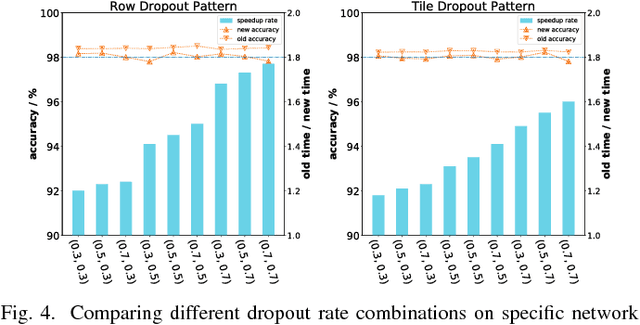
Abstract:The training phases of Deep neural network (DNN) consume enormous processing time and energy. Compression techniques for inference acceleration leveraging the sparsity of DNNs, however, can be hardly used in the training phase. Because the training involves dense matrix-multiplication using GPGPU, which endorse regular and structural data layout. In this paper, we exploit the sparsity of DNN resulting from the random dropout technique to eliminate the unnecessary computation and data access for those dropped neurons or synapses in the training phase. Experiments results on MLP and LSTM on standard benchmarks show that the proposed Approximate Random Dropout can reduce the training time by half on average with ignorable accuracy loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge