Zhenqi He
Category Discovery: An Open-World Perspective
Sep 26, 2025



Abstract:Category discovery (CD) is an emerging open-world learning task, which aims at automatically categorizing unlabelled data containing instances from unseen classes, given some labelled data from seen classes. This task has attracted significant attention over the years and leads to a rich body of literature trying to address the problem from different perspectives. In this survey, we provide a comprehensive review of the literature, and offer detailed analysis and in-depth discussion on different methods. Firstly, we introduce a taxonomy for the literature by considering two base settings, namely novel category discovery (NCD) and generalized category discovery (GCD), and several derived settings that are designed to address the extra challenges in different real-world application scenarios, including continual category discovery, skewed data distribution, federated category discovery, etc. Secondly, for each setting, we offer a detailed analysis of the methods encompassing three fundamental components, representation learning, label assignment, and estimation of class number. Thirdly, we benchmark all the methods and distill key insights showing that large-scale pretrained backbones, hierarchical and auxiliary cues, and curriculum-style training are all beneficial for category discovery, while challenges remain in the design of label assignment, the estimation of class numbers, and scaling to complex multi-object scenarios.Finally, we discuss the key insights from the literature so far and point out promising future research directions. We compile a living survey of the category discovery literature at \href{https://github.com/Visual-AI/Category-Discovery}{https://github.com/Visual-AI/Category-Discovery}.
Hyperbolic Category Discovery
Apr 08, 2025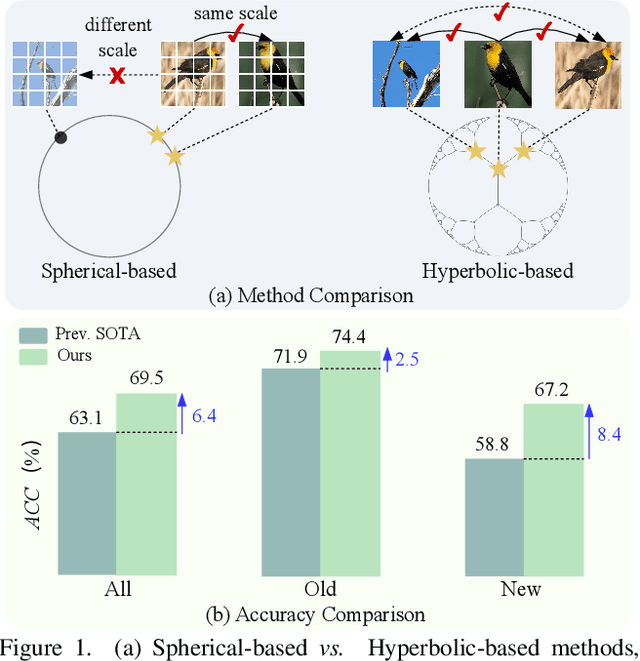
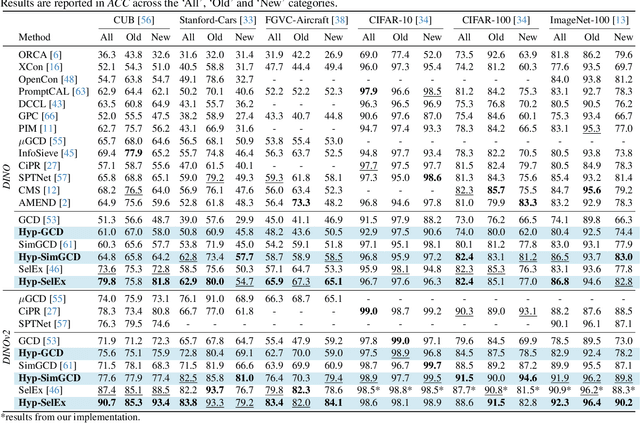
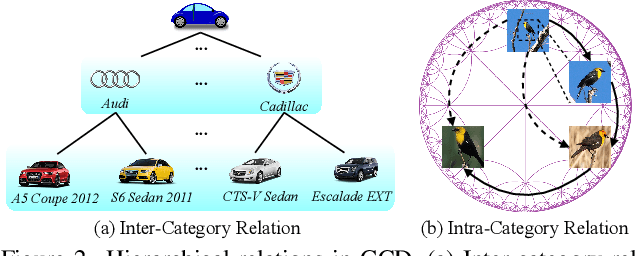
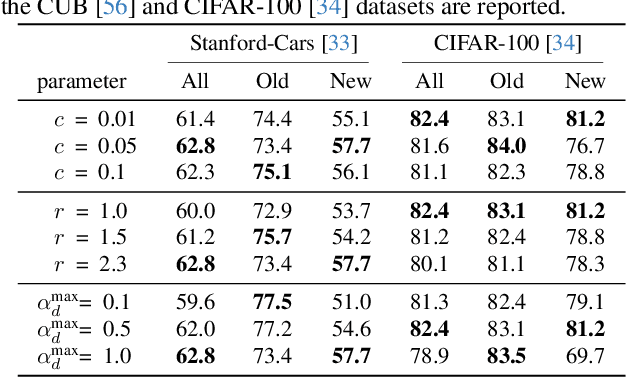
Abstract:Generalized Category Discovery (GCD) is an intriguing open-world problem that has garnered increasing attention. Given a dataset that includes both labelled and unlabelled images, GCD aims to categorize all images in the unlabelled subset, regardless of whether they belong to known or unknown classes. In GCD, the common practice typically involves applying a spherical projection operator at the end of the self-supervised pretrained backbone, operating within Euclidean or spherical space. However, both of these spaces have been shown to be suboptimal for encoding samples that possesses hierarchical structures. In contrast, hyperbolic space exhibits exponential volume growth relative to radius, making it inherently strong at capturing the hierarchical structure of samples from both seen and unseen categories. Therefore, we propose to tackle the category discovery challenge in the hyperbolic space. We introduce HypCD, a simple \underline{Hyp}erbolic framework for learning hierarchy-aware representations and classifiers for generalized \underline{C}ategory \underline{D}iscovery. HypCD first transforms the Euclidean embedding space of the backbone network into hyperbolic space, facilitating subsequent representation and classification learning by considering both hyperbolic distance and the angle between samples. This approach is particularly helpful for knowledge transfer from known to unknown categories in GCD. We thoroughly evaluate HypCD on public GCD benchmarks, by applying it to various baseline and state-of-the-art methods, consistently achieving significant improvements.
Towards Hierarchical Multi-Step Reward Models for Enhanced Reasoning in Large Language Models
Mar 19, 2025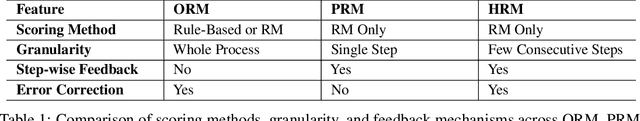
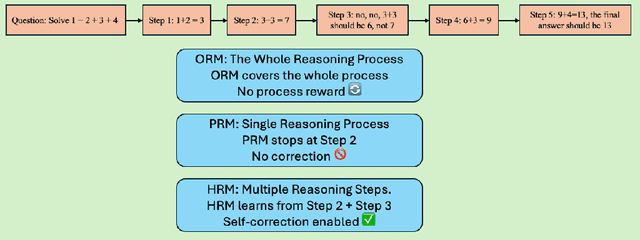
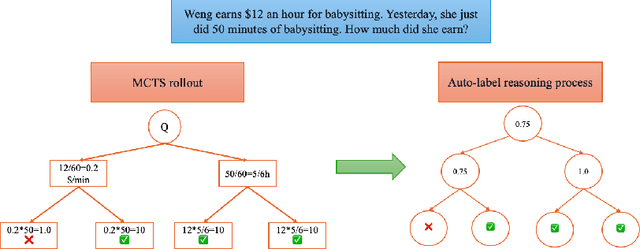
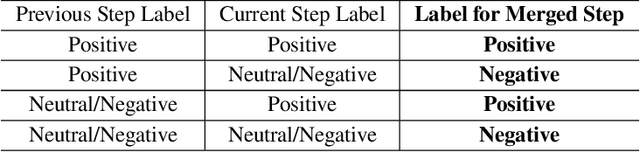
Abstract:Recent studies show that Large Language Models (LLMs) achieve strong reasoning capabilities through supervised fine-tuning or reinforcement learning. However, a key approach, the Process Reward Model (PRM), suffers from reward hacking, making it unreliable in identifying the best intermediate steps. In this paper, we propose a novel reward model approach, Hierarchical Reward Model (HRM), which evaluates both individual and consecutive reasoning steps from fine-grained and coarse-grained level. HRM performs better in assessing reasoning coherence and self-reflection, particularly when the previous reasoning step is incorrect. Furthermore, to address the inefficiency of autonomous generating PRM training data via Monte Carlo Tree Search (MCTS), we introduce a lightweight and effective data augmentation strategy called Hierarchical Node Compression (HNC) based on node merging (combining two consecutive reasoning steps into one step) in the tree structure. This approach diversifies MCTS results for HRM with negligible computational overhead, enhancing label robustness by introducing noise. Empirical results on the PRM800K dataset demonstrate that HRM, in conjunction with HNC, achieves superior stability and reliability in evaluation compared to PRM. Furthermore, cross-domain evaluations on MATH500 and GSM8K confirm HRM's superior generalization and robustness across diverse reasoning tasks. The code for all experiments will be released at https: //github.com/tengwang0318/hierarchial_reward_model.
BPP-Search: Enhancing Tree of Thought Reasoning for Mathematical Modeling Problem Solving
Nov 26, 2024



Abstract:LLMs exhibit advanced reasoning capabilities, offering the potential to transform natural language questions into mathematical models. However, existing open-source operations research datasets lack detailed annotations of the modeling process, such as variable definitions, focusing solely on objective values, which hinders reinforcement learning applications. To address this, we release the StructuredOR dataset, annotated with comprehensive labels that capture the complete mathematical modeling process. We further propose BPP-Search, a algorithm that integrates reinforcement learning into a tree-of-thought structure using Beam search, a Process reward model, and a pairwise Preference algorithm. This approach enables efficient exploration of tree structures, avoiding exhaustive search while improving accuracy. Extensive experiments on StructuredOR, NL4OPT, and MAMO-ComplexLP datasets show that BPP-Search significantly outperforms state-of-the-art methods, including Chain-of-Thought, Self-Consistency, and Tree-of-Thought. In tree-based reasoning, BPP-Search also surpasses Process Reward Model combined with Greedy or Beam Search, demonstrating superior accuracy and efficiency, and enabling faster retrieval of correct solutions.
Large Language Models are Good Multi-lingual Learners : When LLMs Meet Cross-lingual Prompts
Sep 17, 2024



Abstract:With the advent of Large Language Models (LLMs), generating rule-based data for real-world applications has become more accessible. Due to the inherent ambiguity of natural language and the complexity of rule sets, especially in long contexts, LLMs often struggle to follow all specified rules, frequently omitting at least one. To enhance the reasoning and understanding of LLMs on long and complex contexts, we propose a novel prompting strategy Multi-Lingual Prompt, namely MLPrompt, which automatically translates the error-prone rule that an LLM struggles to follow into another language, thus drawing greater attention to it. Experimental results on public datasets across various tasks have shown MLPrompt can outperform state-of-the-art prompting methods such as Chain of Thought, Tree of Thought, and Self-Consistency. Additionally, we introduce a framework integrating MLPrompt with an auto-checking mechanism for structured data generation, with a specific case study in text-to-MIP instances. Further, we extend the proposed framework for text-to-SQL to demonstrate its generation ability towards structured data synthesis.
LatentArtiFusion: An Effective and Efficient Histological Artifacts Restoration Framework
Jul 29, 2024



Abstract:Histological artifacts pose challenges for both pathologists and Computer-Aided Diagnosis (CAD) systems, leading to errors in analysis. Current approaches for histological artifact restoration, based on Generative Adversarial Networks (GANs) and pixel-level Diffusion Models, suffer from performance limitations and computational inefficiencies. In this paper, we propose a novel framework, LatentArtiFusion, which leverages the latent diffusion model (LDM) to reconstruct histological artifacts with high performance and computational efficiency. Unlike traditional pixel-level diffusion frameworks, LatentArtiFusion executes the restoration process in a lower-dimensional latent space, significantly improving computational efficiency. Moreover, we introduce a novel regional artifact reconstruction algorithm in latent space to prevent mistransfer in non-artifact regions, distinguishing our approach from GAN-based methods. Through extensive experiments on real-world histology datasets, LatentArtiFusion demonstrates remarkable speed, outperforming state-of-the-art pixel-level diffusion frameworks by more than 30X. It also consistently surpasses GAN-based methods by at least 5% across multiple evaluation metrics. Furthermore, we evaluate the effectiveness of our proposed framework in downstream tissue classification tasks, showcasing its practical utility. Code is available at https://github.com/bugs-creator/LatentArtiFusion.
Artifact Restoration in Histology Images with Diffusion Probabilistic Models
Jul 26, 2023



Abstract:Histological whole slide images (WSIs) can be usually compromised by artifacts, such as tissue folding and bubbles, which will increase the examination difficulty for both pathologists and Computer-Aided Diagnosis (CAD) systems. Existing approaches to restoring artifact images are confined to Generative Adversarial Networks (GANs), where the restoration process is formulated as an image-to-image transfer. Those methods are prone to suffer from mode collapse and unexpected mistransfer in the stain style, leading to unsatisfied and unrealistic restored images. Innovatively, we make the first attempt at a denoising diffusion probabilistic model for histological artifact restoration, namely ArtiFusion.Specifically, ArtiFusion formulates the artifact region restoration as a gradual denoising process, and its training relies solely on artifact-free images to simplify the training complexity.Furthermore, to capture local-global correlations in the regional artifact restoration, a novel Swin-Transformer denoising architecture is designed, along with a time token scheme. Our extensive evaluations demonstrate the effectiveness of ArtiFusion as a pre-processing method for histology analysis, which can successfully preserve the tissue structures and stain style in artifact-free regions during the restoration. Code is available at https://github.com/zhenqi-he/ArtiFusion.
TransNuSeg: A Lightweight Multi-Task Transformer for Nuclei Segmentation
Jul 16, 2023Abstract:Nuclei appear small in size, yet, in real clinical practice, the global spatial information and correlation of the color or brightness contrast between nuclei and background, have been considered a crucial component for accurate nuclei segmentation. However, the field of automatic nuclei segmentation is dominated by Convolutional Neural Networks (CNNs), meanwhile, the potential of the recently prevalent Transformers has not been fully explored, which is powerful in capturing local-global correlations. To this end, we make the first attempt at a pure Transformer framework for nuclei segmentation, called TransNuSeg. Different from prior work, we decouple the challenging nuclei segmentation task into an intrinsic multi-task learning task, where a tri-decoder structure is employed for nuclei instance, nuclei edge, and clustered edge segmentation respectively. To eliminate the divergent predictions from different branches in previous work, a novel self distillation loss is introduced to explicitly impose consistency regulation between branches. Moreover, to formulate the high correlation between branches and also reduce the number of parameters, an efficient attention sharing scheme is proposed by partially sharing the self-attention heads amongst the tri-decoders. Finally, a token MLP bottleneck replaces the over-parameterized Transformer bottleneck for a further reduction in model complexity. Experiments on two datasets of different modalities, including MoNuSeg have shown that our methods can outperform state-of-the-art counterparts such as CA2.5-Net by 2-3% Dice with 30% fewer parameters. In conclusion, TransNuSeg confirms the strength of Transformer in the context of nuclei segmentation, which thus can serve as an efficient solution for real clinical practice. Code is available at https://github.com/zhenqi-he/transnuseg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge