Heimo Müller
World Models in Artificial Intelligence: Sensing, Learning, and Reasoning Like a Child
Mar 19, 2025Abstract:World Models help Artificial Intelligence (AI) predict outcomes, reason about its environment, and guide decision-making. While widely used in reinforcement learning, they lack the structured, adaptive representations that even young children intuitively develop. Advancing beyond pattern recognition requires dynamic, interpretable frameworks inspired by Piaget's cognitive development theory. We highlight six key research areas -- physics-informed learning, neurosymbolic learning, continual learning, causal inference, human-in-the-loop AI, and responsible AI -- as essential for enabling true reasoning in AI. By integrating statistical learning with advances in these areas, AI can evolve from pattern recognition to genuine understanding, adaptation and reasoning capabilities.
Diffusion-based generation of Histopathological Whole Slide Images at a Gigapixel scale
Nov 14, 2023Abstract:We present a novel diffusion-based approach to generate synthetic histopathological Whole Slide Images (WSIs) at an unprecedented gigapixel scale. Synthetic WSIs have many potential applications: They can augment training datasets to enhance the performance of many computational pathology applications. They allow the creation of synthesized copies of datasets that can be shared without violating privacy regulations. Or they can facilitate learning representations of WSIs without requiring data annotations. Despite this variety of applications, no existing deep-learning-based method generates WSIs at their typically high resolutions. Mainly due to the high computational complexity. Therefore, we propose a novel coarse-to-fine sampling scheme to tackle image generation of high-resolution WSIs. In this scheme, we increase the resolution of an initial low-resolution image to a high-resolution WSI. Particularly, a diffusion model sequentially adds fine details to images and increases their resolution. In our experiments, we train our method with WSIs from the TCGA-BRCA dataset. Additionally to quantitative evaluations, we also performed a user study with pathologists. The study results suggest that our generated WSIs resemble the structure of real WSIs.
Ethical ChatGPT: Concerns, Challenges, and Commandments
May 18, 2023Abstract:Large language models, e.g. ChatGPT are currently contributing enormously to make artificial intelligence even more popular, especially among the general population. However, such chatbot models were developed as tools to support natural language communication between humans. Problematically, it is very much a ``statistical correlation machine" (correlation instead of causality) and there are indeed ethical concerns associated with the use of AI language models such as ChatGPT, such as Bias, Privacy, and Abuse. This paper highlights specific ethical concerns on ChatGPT and articulates key challenges when ChatGPT is used in various applications. Practical commandments for different stakeholders of ChatGPT are also proposed that can serve as checklist guidelines for those applying ChatGPT in their applications. These commandment examples are expected to motivate the ethical use of ChatGPT.
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022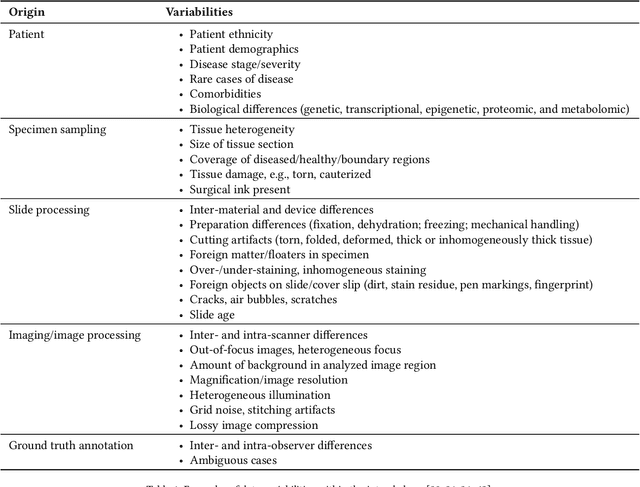
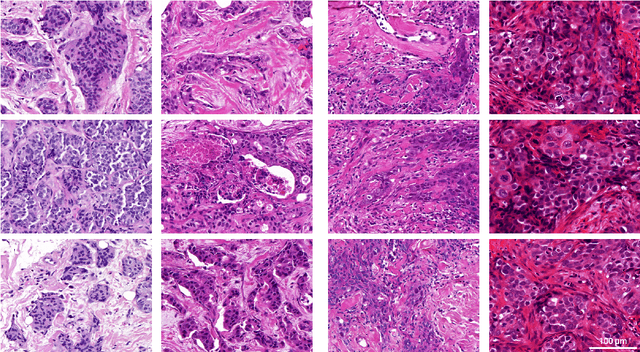
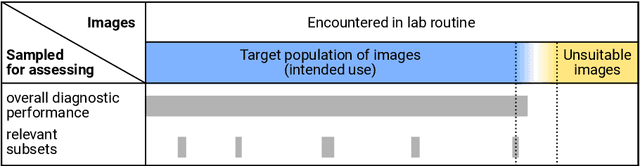
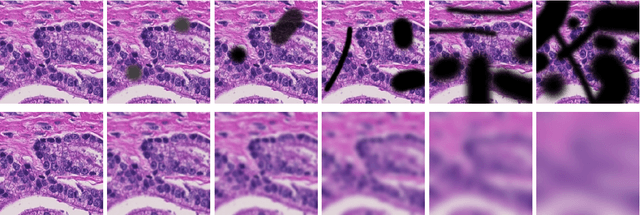
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
The FeatureCloud AI Store for Federated Learning in Biomedicine and Beyond
May 12, 2021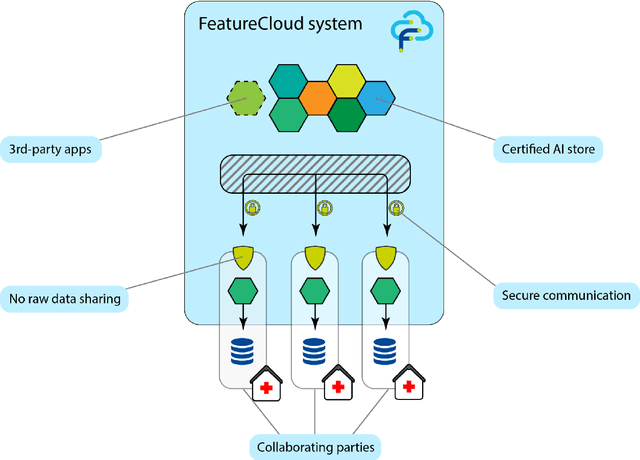
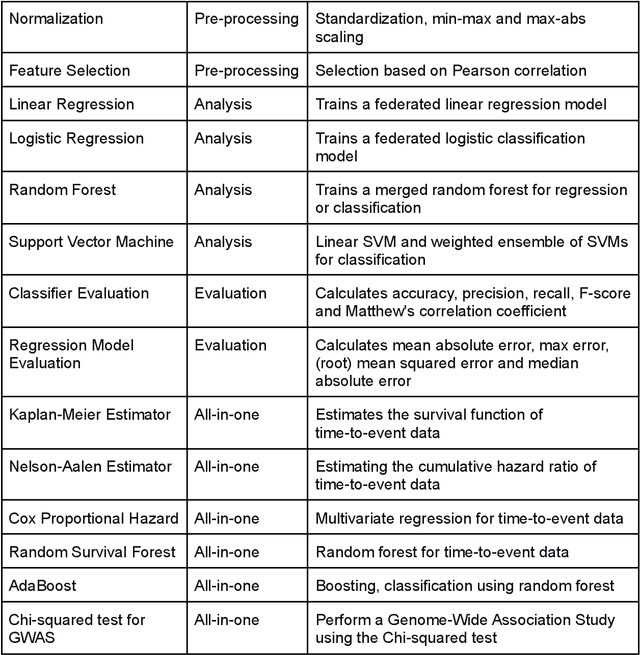
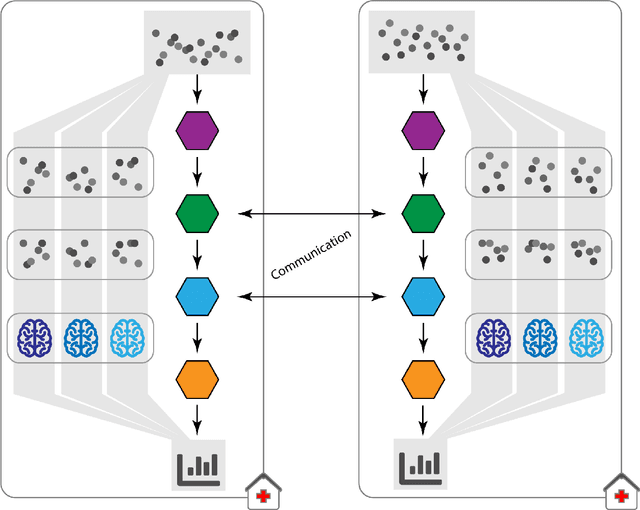
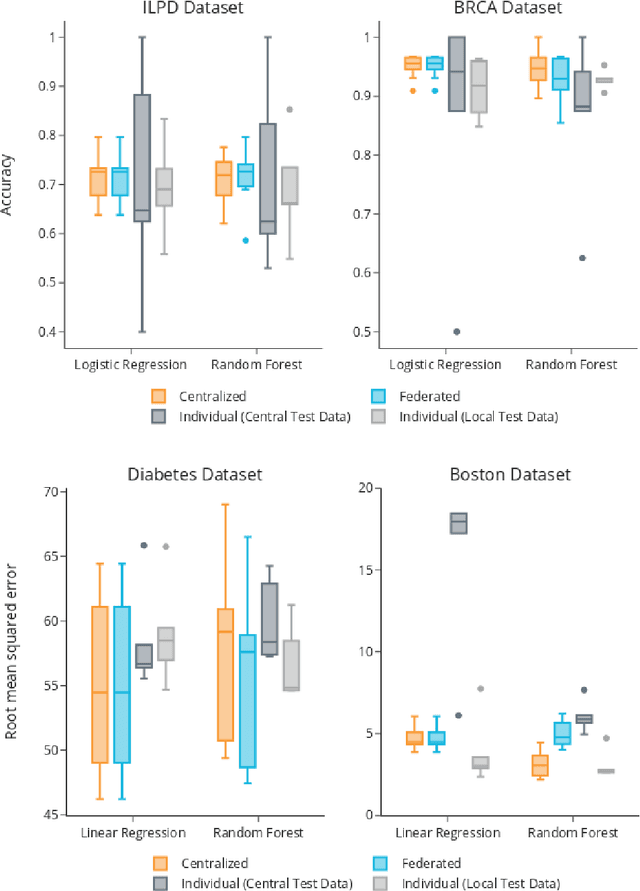
Abstract:Machine Learning (ML) and Artificial Intelligence (AI) have shown promising results in many areas and are driven by the increasing amount of available data. However, this data is often distributed across different institutions and cannot be shared due to privacy concerns. Privacy-preserving methods, such as Federated Learning (FL), allow for training ML models without sharing sensitive data, but their implementation is time-consuming and requires advanced programming skills. Here, we present the FeatureCloud AI Store for FL as an all-in-one platform for biomedical research and other applications. It removes large parts of this complexity for developers and end-users by providing an extensible AI Store with a collection of ready-to-use apps. We show that the federated apps produce similar results to centralized ML, scale well for a typical number of collaborators and can be combined with Secure Multiparty Computation (SMPC), thereby making FL algorithms safely and easily applicable in biomedical and clinical environments.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020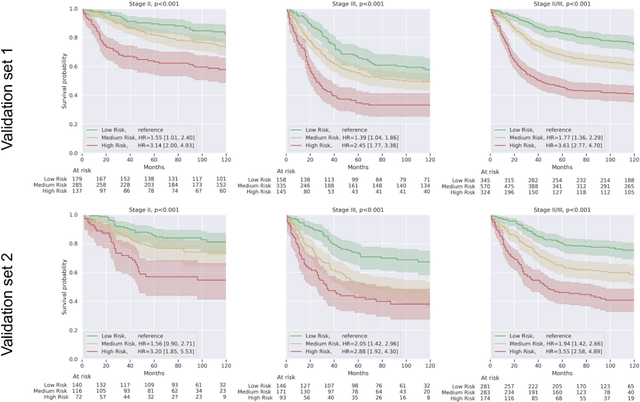
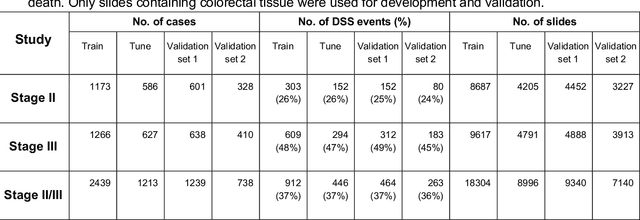
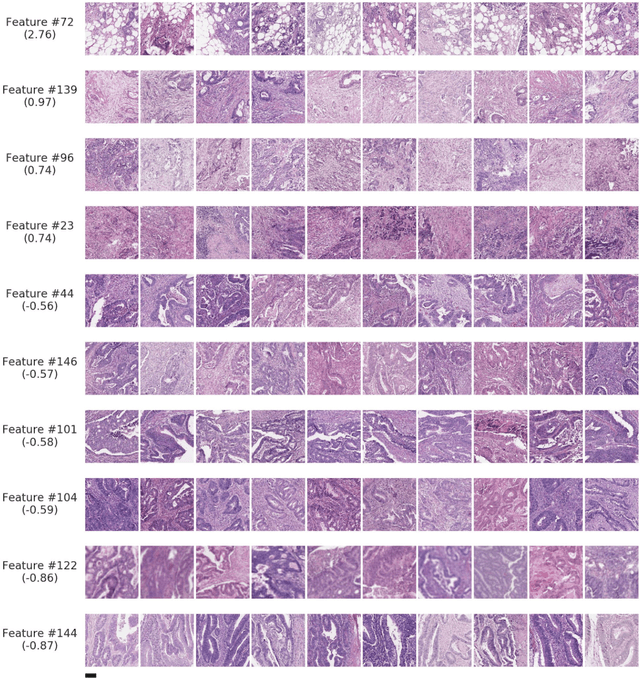
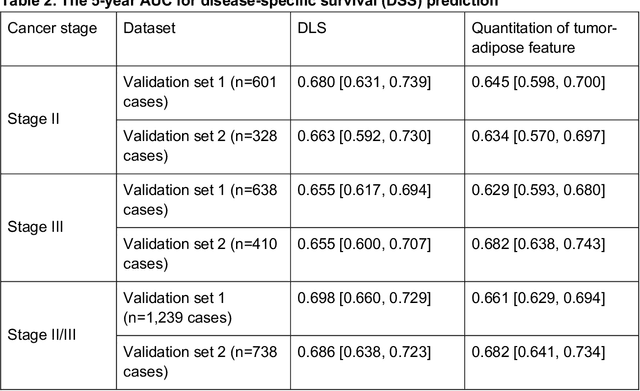
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
Measuring the Quality of Explanations: The System Causability Scale (SCS). Comparing Human and Machine Explanations
Dec 19, 2019

Abstract:Recent success in Artificial Intelligence (AI) and Machine Learning (ML) allow problem solving automatically without any human intervention. Autonomous approaches can be very convenient. However, in certain domains, e.g., in the medical domain, it is necessary to enable a domain expert to understand, why an algorithm came up with a certain result. Consequently, the field of Explainable AI (xAI) rapidly gained interest worldwide in various domains, particularly in medicine. Explainable AI studies transparency and traceability of opaque AI/ML and there are already a huge variety of methods. For example with layer-wise relevance propagation relevant parts of inputs to, and representations in, a neural network which caused a result, can be highlighted. This is a first important step to ensure that end users, e.g., medical professionals, assume responsibility for decision making with AI/ML and of interest to professionals and regulators. Interactive ML adds the component of human expertise to AI/ML processes by enabling them to re-enact and retrace AI/ML results, e.g. let them check it for plausibility. This requires new human-AI interfaces for explainable AI. In order to build effective and efficient interactive human-AI interfaces we have to deal with the question of how to evaluate the quality of explanations given by an explainable AI system. In this paper we introduce our System Causability Scale (SCS) to measure the quality of explanations. It is based on our notion of Causability (Holzinger et al., 2019) combined with concepts adapted from a widely accepted usability scale.
Towards the Augmented Pathologist: Challenges of Explainable-AI in Digital Pathology
Dec 18, 2017



Abstract:Digital pathology is not only one of the most promising fields of diagnostic medicine, but at the same time a hot topic for fundamental research. Digital pathology is not just the transfer of histopathological slides into digital representations. The combination of different data sources (images, patient records, and *omics data) together with current advances in artificial intelligence/machine learning enable to make novel information accessible and quantifiable to a human expert, which is not yet available and not exploited in current medical settings. The grand goal is to reach a level of usable intelligence to understand the data in the context of an application task, thereby making machine decisions transparent, interpretable and explainable. The foundation of such an "augmented pathologist" needs an integrated approach: While machine learning algorithms require many thousands of training examples, a human expert is often confronted with only a few data points. Interestingly, humans can learn from such few examples and are able to instantly interpret complex patterns. Consequently, the grand goal is to combine the possibilities of artificial intelligence with human intelligence and to find a well-suited balance between them to enable what neither of them could do on their own. This can raise the quality of education, diagnosis, prognosis and prediction of cancer and other diseases. In this paper we describe some (incomplete) research issues which we believe should be addressed in an integrated and concerted effort for paving the way towards the augmented pathologist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge