Lars Ole Schwen
Digitization of Pathology Labs: A Review of Lessons Learned
Jun 07, 2023


Abstract:Pathology laboratories are increasingly using digital workflows. This has the potential of increasing lab efficiency, but the digitization process also involves major challenges. Several reports have been published describing the individual experiences of specific laboratories with the digitization process. However, a comprehensive overview of the lessons learned is still lacking. We provide an overview of the lessons learned for different aspects of the digitization process, including digital case management, digital slide reading, and computer-aided slide reading. We also cover metrics used for monitoring performance and pitfalls and corresponding values observed in practice. The overview is intended to help pathologists, IT decision-makers, and administrators to benefit from the experiences of others and to implement the digitization process in an optimal way to make their own laboratory future-proof.
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022
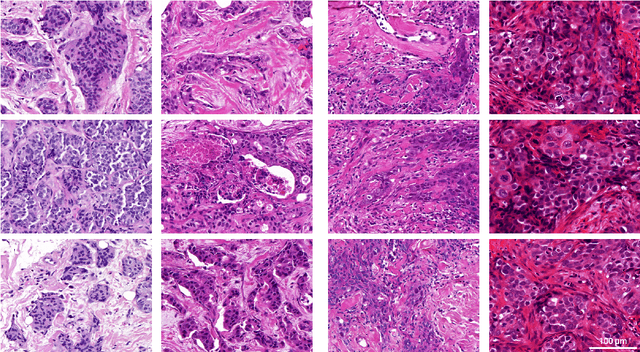
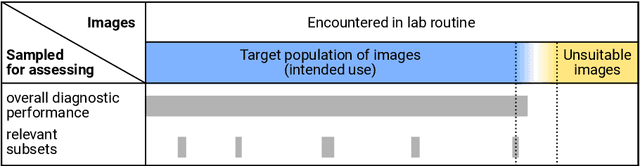
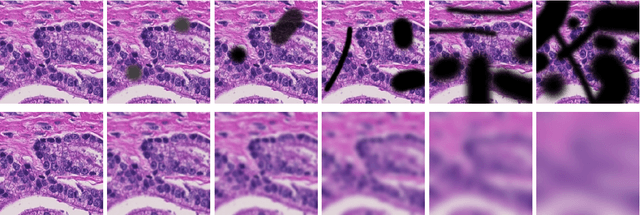
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
Evaluating Generic Auto-ML Tools for Computational Pathology
Dec 07, 2021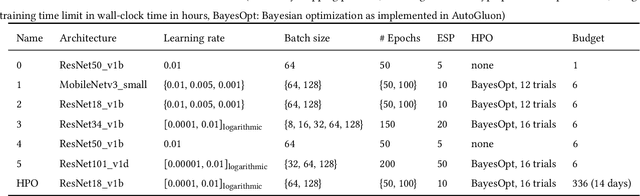
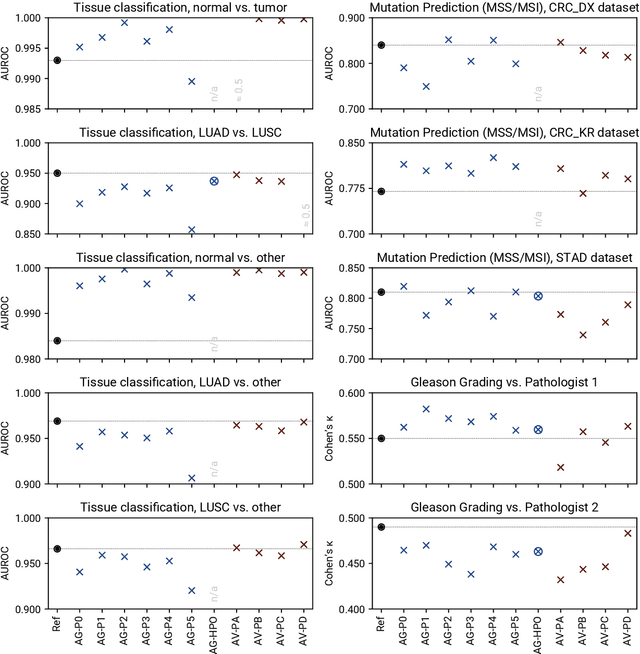

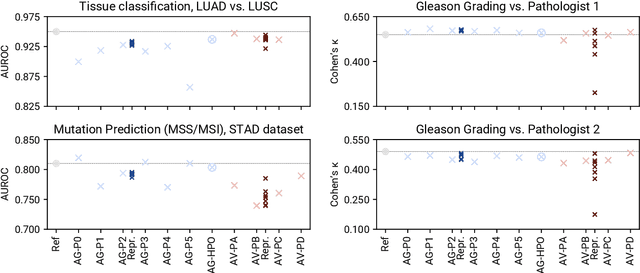
Abstract:Image analysis tasks in computational pathology are commonly solved using convolutional neural networks (CNNs). The selection of a suitable CNN architecture and hyperparameters is usually done through exploratory iterative optimization, which is computationally expensive and requires substantial manual work. The goal of this article is to evaluate how generic tools for neural network architecture search and hyperparameter optimization perform for common use cases in computational pathology. For this purpose, we evaluated one on-premises and one cloud-based tool for three different classification tasks for histological images: tissue classification, mutation prediction, and grading. We found that the default CNN architectures and parameterizations of the evaluated AutoML tools already yielded classification performance on par with the original publications. Hyperparameter optimization for these tasks did not substantially improve performance, despite the additional computational effort. However, performance varied substantially between classifiers obtained from individual AutoML runs due to non-deterministic effects. Generic CNN architectures and AutoML tools could thus be a viable alternative to manually optimizing CNN architectures and parametrizations. This would allow developers of software solutions for computational pathology to focus efforts on harder-to-automate tasks such as data curation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge