Falk Zakrzewski
Recommendations on test datasets for evaluating AI solutions in pathology
Apr 21, 2022
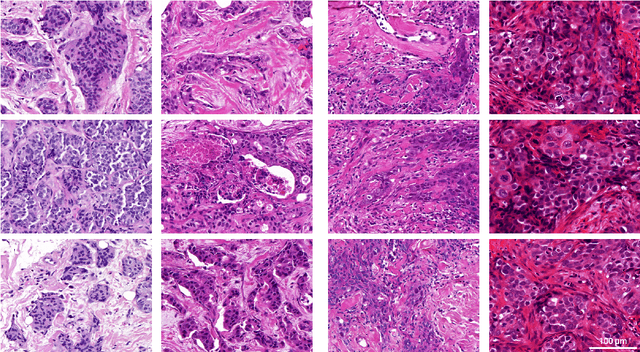
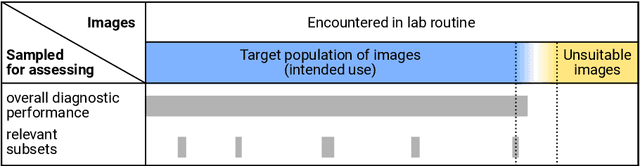
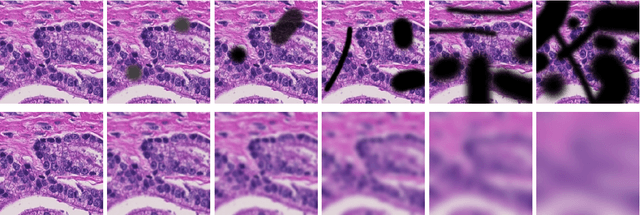
Abstract:Artificial intelligence (AI) solutions that automatically extract information from digital histology images have shown great promise for improving pathological diagnosis. Prior to routine use, it is important to evaluate their predictive performance and obtain regulatory approval. This assessment requires appropriate test datasets. However, compiling such datasets is challenging and specific recommendations are missing. A committee of various stakeholders, including commercial AI developers, pathologists, and researchers, discussed key aspects and conducted extensive literature reviews on test datasets in pathology. Here, we summarize the results and derive general recommendations for the collection of test datasets. We address several questions: Which and how many images are needed? How to deal with low-prevalence subsets? How can potential bias be detected? How should datasets be reported? What are the regulatory requirements in different countries? The recommendations are intended to help AI developers demonstrate the utility of their products and to help regulatory agencies and end users verify reported performance measures. Further research is needed to formulate criteria for sufficiently representative test datasets so that AI solutions can operate with less user intervention and better support diagnostic workflows in the future.
An interpretable automated detection system for FISH-based HER2 oncogene amplification testing in histo-pathological routine images of breast and gastric cancer diagnostics
May 25, 2020
Abstract:Histo-pathological diagnostics are an inherent part of the everyday work but are particularly laborious and associated with time-consuming manual analysis of image data. In order to cope with the increasing diagnostic case numbers due to the current growth and demographic change of the global population and the progress in personalized medicine, pathologists ask for assistance. Profiting from digital pathology and the use of artificial intelligence, individual solutions can be offered (e.g. detect labeled cancer tissue sections). The testing of the human epidermal growth factor receptor 2 (HER2) oncogene amplification status via fluorescence in situ hybridization (FISH) is recommended for breast and gastric cancer diagnostics and is regularly performed at clinics. Here, we develop an interpretable, deep learning (DL)-based pipeline which automates the evaluation of FISH images with respect to HER2 gene amplification testing. It mimics the pathological assessment and relies on the detection and localization of interphase nuclei based on instance segmentation networks. Furthermore, it localizes and classifies fluorescence signals within each nucleus with the help of image classification and object detection convolutional neural networks (CNNs). Finally, the pipeline classifies the whole image regarding its HER2 amplification status. The visualization of pixels on which the networks' decision occurs, complements an essential part to enable interpretability by pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge