Ingo Roeder
Fair Distributed Machine Learning with Imbalanced Data as a Stackelberg Evolutionary Game
Dec 20, 2024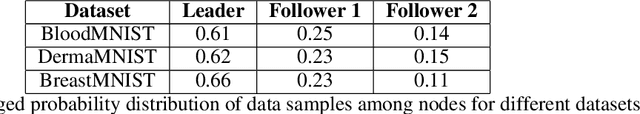
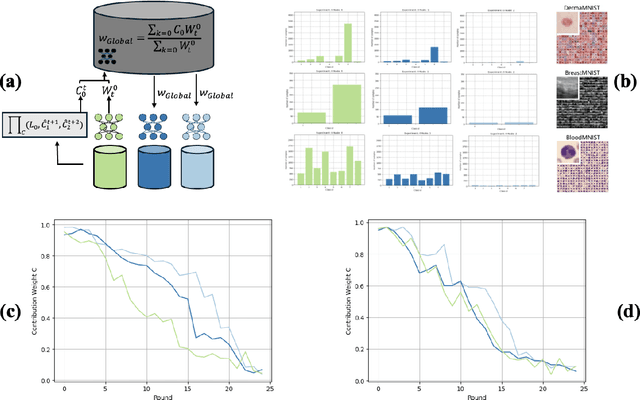
Abstract:Decentralised learning enables the training of deep learning algorithms without centralising data sets, resulting in benefits such as improved data privacy, operational efficiency and the fostering of data ownership policies. However, significant data imbalances pose a challenge in this framework. Participants with smaller datasets in distributed learning environments often achieve poorer results than participants with larger datasets. Data imbalances are particularly pronounced in medical fields and are caused by different patient populations, technological inequalities and divergent data collection practices. In this paper, we consider distributed learning as an Stackelberg evolutionary game. We present two algorithms for setting the weights of each node's contribution to the global model in each training round: the Deterministic Stackelberg Weighting Model (DSWM) and the Adaptive Stackelberg Weighting Model (ASWM). We use three medical datasets to highlight the impact of dynamic weighting on underrepresented nodes in distributed learning. Our results show that the ASWM significantly favours underrepresented nodes by improving their performance by 2.713% in AUC. Meanwhile, nodes with larger datasets experience only a modest average performance decrease of 0.441%.
Quality control for more reliable integration of deep learning-based image segmentation into medical workflows
Dec 06, 2021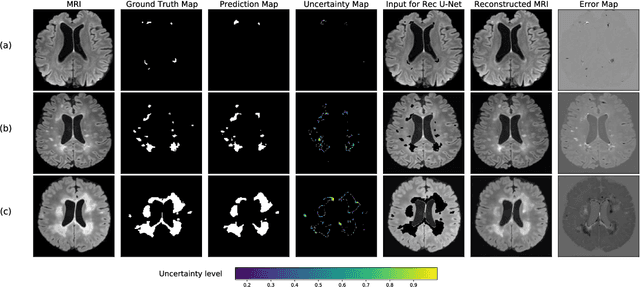
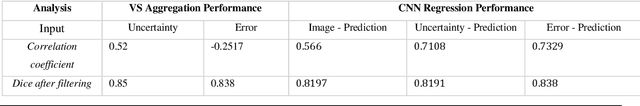
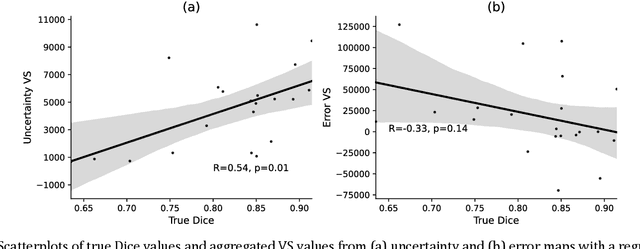
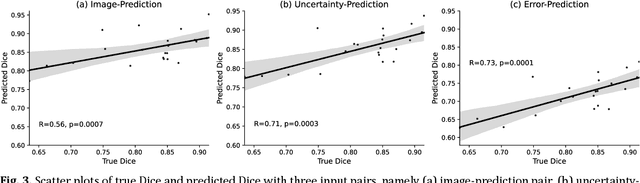
Abstract:Machine learning algorithms underpin modern diagnostic-aiding software, which has proved valuable in clinical practice, particularly in radiology. However, inaccuracies, mainly due to the limited availability of clinical samples for training these algorithms, hamper their wider applicability, acceptance, and recognition amongst clinicians. We present an analysis of state-of-the-art automatic quality control (QC) approaches that can be implemented within these algorithms to estimate the certainty of their outputs. We validated the most promising approaches on a brain image segmentation task identifying white matter hyperintensities (WMH) in magnetic resonance imaging data. WMH are a correlate of small vessel disease common in mid-to-late adulthood and are particularly challenging to segment due to their varied size, and distributional patterns. Our results show that the aggregation of uncertainty and Dice prediction were most effective in failure detection for this task. Both methods independently improved mean Dice from 0.82 to 0.84. Our work reveals how QC methods can help to detect failed segmentation cases and therefore make automatic segmentation more reliable and suitable for clinical practice.
A comparative study of semi- and self-supervised semantic segmentation of biomedical microscopy data
Nov 23, 2020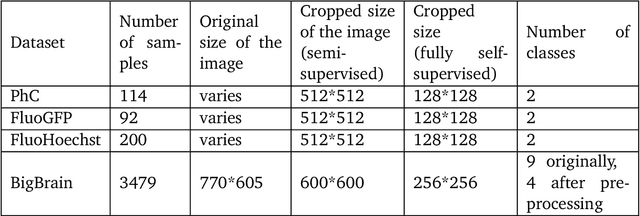

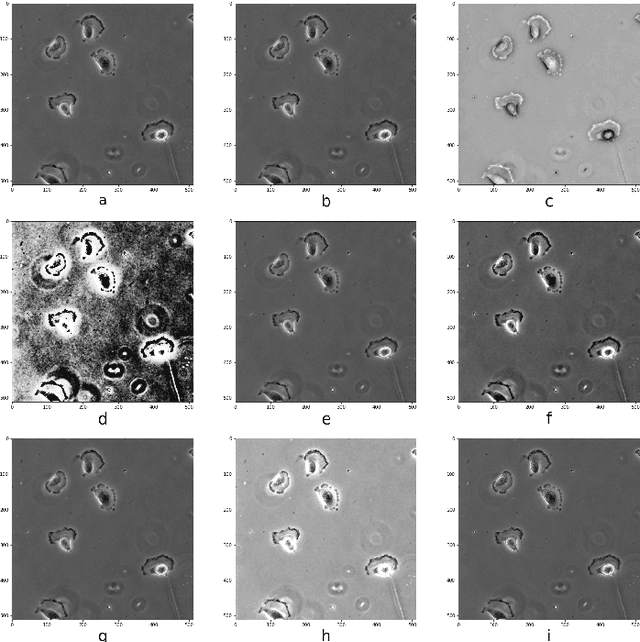
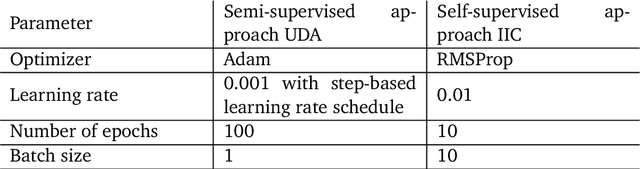
Abstract:In recent years, Convolutional Neural Networks (CNNs) have become the state-of-the-art method for biomedical image analysis. However, these networks are usually trained in a supervised manner, requiring large amounts of labelled training data. These labelled data sets are often difficult to acquire in the biomedical domain. In this work, we validate alternative ways to train CNNs with fewer labels for biomedical image segmentation using. We adapt two semi- and self-supervised image classification methods and analyse their performance for semantic segmentation of biomedical microscopy images.
An interpretable automated detection system for FISH-based HER2 oncogene amplification testing in histo-pathological routine images of breast and gastric cancer diagnostics
May 25, 2020
Abstract:Histo-pathological diagnostics are an inherent part of the everyday work but are particularly laborious and associated with time-consuming manual analysis of image data. In order to cope with the increasing diagnostic case numbers due to the current growth and demographic change of the global population and the progress in personalized medicine, pathologists ask for assistance. Profiting from digital pathology and the use of artificial intelligence, individual solutions can be offered (e.g. detect labeled cancer tissue sections). The testing of the human epidermal growth factor receptor 2 (HER2) oncogene amplification status via fluorescence in situ hybridization (FISH) is recommended for breast and gastric cancer diagnostics and is regularly performed at clinics. Here, we develop an interpretable, deep learning (DL)-based pipeline which automates the evaluation of FISH images with respect to HER2 gene amplification testing. It mimics the pathological assessment and relies on the detection and localization of interphase nuclei based on instance segmentation networks. Furthermore, it localizes and classifies fluorescence signals within each nucleus with the help of image classification and object detection convolutional neural networks (CNNs). Finally, the pipeline classifies the whole image regarding its HER2 amplification status. The visualization of pixels on which the networks' decision occurs, complements an essential part to enable interpretability by pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge