Eran Segal
Weizmann Institute of Science, Rehovot, Israel
MoHETS: Long-term Time Series Forecasting with Mixture-of-Heterogeneous-Experts
Jan 29, 2026Abstract:Real-world multivariate time series can exhibit intricate multi-scale structures, including global trends, local periodicities, and non-stationary regimes, which makes long-horizon forecasting challenging. Although sparse Mixture-of-Experts (MoE) approaches improve scalability and specialization, they typically rely on homogeneous MLP experts that poorly capture the diverse temporal dynamics of time series data. We address these limitations with MoHETS, an encoder-only Transformer that integrates sparse Mixture-of-Heterogeneous-Experts (MoHE) layers. MoHE routes temporal patches to a small subset of expert networks, combining a shared depthwise-convolution expert for sequence-level continuity with routed Fourier-based experts for patch-level periodic structures. MoHETS further improves robustness to non-stationary dynamics by incorporating exogenous information via cross-attention over covariate patch embeddings. Finally, we replace parameter-heavy linear projection heads with a lightweight convolutional patch decoder, improving parameter efficiency, reducing training instability, and allowing a single model to generalize across arbitrary forecast horizons. We validate across seven multivariate benchmarks and multiple horizons, with MoHETS consistently achieving state-of-the-art performance, reducing the average MSE by $12\%$ compared to strong recent baselines, demonstrating effective heterogeneous specialization for long-term forecasting.
Seg-MoE: Multi-Resolution Segment-wise Mixture-of-Experts for Time Series Forecasting Transformers
Jan 29, 2026Abstract:Transformer-based models have recently made significant advances in accurate time-series forecasting, but even these architectures struggle to scale efficiently while capturing long-term temporal dynamics. Mixture-of-Experts (MoE) layers are a proven solution to scaling problems in natural language processing. However, existing MoE approaches for time-series forecasting rely on token-wise routing mechanisms, which may fail to exploit the natural locality and continuity of temporal data. In this work, we introduce Seg-MoE, a sparse MoE design that routes and processes contiguous time-step segments rather than making independent expert decisions. Token segments allow each expert to model intra-segment interactions directly, naturally aligning with inherent temporal patterns. We integrate Seg-MoE layers into a time-series Transformer and evaluate it on multiple multivariate long-term forecasting benchmarks. Seg-MoE consistently achieves state-of-the-art forecasting accuracy across almost all prediction horizons, outperforming both dense Transformers and prior token-wise MoE models. Comprehensive ablation studies confirm that segment-level routing is the key factor driving these gains. Our results show that aligning the MoE routing granularity with the inherent structure of time series provides a powerful, yet previously underexplored, inductive bias, opening new avenues for conditionally sparse architectures in sequential data modeling.
Phenome-Wide Multi-Omics Integration Uncovers Distinct Archetypes of Human Aging
Oct 14, 2025Abstract:Aging is a highly complex and heterogeneous process that progresses at different rates across individuals, making biological age (BA) a more accurate indicator of physiological decline than chronological age. While previous studies have built aging clocks using single-omics data, they often fail to capture the full molecular complexity of human aging. In this work, we leveraged the Human Phenotype Project, a large-scale cohort of 12,000 adults aged 30--70 years, with extensive longitudinal profiling that includes clinical, behavioral, environmental, and multi-omics datasets -- spanning transcriptomics, lipidomics, metabolomics, and the microbiome. By employing advanced machine learning frameworks capable of modeling nonlinear biological dynamics, we developed and rigorously validated a multi-omics aging clock that robustly predicts diverse health outcomes and future disease risk. Unsupervised clustering of the integrated molecular profiles from multi-omics uncovered distinct biological subtypes of aging, revealing striking heterogeneity in aging trajectories and pinpointing pathway-specific alterations associated with different aging patterns. These findings demonstrate the power of multi-omics integration to decode the molecular landscape of aging and lay the groundwork for personalized healthspan monitoring and precision strategies to prevent age-related diseases.
HPP-Voice: A Large-Scale Evaluation of Speech Embeddings for Multi-Phenotypic Classification
May 22, 2025Abstract:Human speech contains paralinguistic cues that reflect a speaker's physiological and neurological state, potentially enabling non-invasive detection of various medical phenotypes. We introduce the Human Phenotype Project Voice corpus (HPP-Voice): a dataset of 7,188 recordings in which Hebrew-speaking adults count for 30 seconds, with each speaker linked to up to 15 potentially voice-related phenotypes spanning respiratory, sleep, mental health, metabolic, immune, and neurological conditions. We present a systematic comparison of 14 modern speech embedding models, where modern speech embeddings from these 30-second counting tasks outperform MFCCs and demographics for downstream health condition classifications. We found that embedding learned from a speaker identification model can predict objectively measured moderate to severe sleep apnea in males with an AUC of 0.64 $\pm$ 0.03, while MFCC and demographic features led to AUCs of 0.56 $\pm$ 0.02 and 0.57 $\pm$ 0.02, respectively. Additionally, our results reveal gender-specific patterns in model effectiveness across different medical domains. For males, speaker identification and diarization models consistently outperformed speech foundation models for respiratory conditions (e.g., asthma: 0.61 $\pm$ 0.03 vs. 0.56 $\pm$ 0.02) and sleep-related conditions (insomnia: 0.65 $\pm$ 0.04 vs. 0.59 $\pm$ 0.05). For females, speaker diarization models performed best for smoking status (0.61 $\pm$ 0.02 vs 0.55 $\pm$ 0.02), while Hebrew-specific models performed best (0.59 $\pm$ 0.02 vs. 0.58 $\pm$ 0.02) in classifying anxiety compared to speech foundation models. Our findings provide evidence that a simple counting task can support large-scale, multi-phenotypic voice screening and highlight which embedding families generalize best to specific conditions, insights that can guide future vocal biomarker research and clinical deployment.
Improving Diseases Predictions Utilizing External Bio-Banks
Mar 30, 2025



Abstract:Machine learning has been successfully used in critical domains, such as medicine. However, extracting meaningful insights from biomedical data is often constrained by the lack of their available disease labels. In this research, we demonstrate how machine learning can be leveraged to enhance explainability and uncover biologically meaningful associations, even when predictive improvements in disease modeling are limited. We train LightGBM models from scratch on our dataset (10K) to impute metabolomics features and apply them to the UK Biobank (UKBB) for downstream analysis. The imputed metabolomics features are then used in survival analysis to assess their impact on disease-related risk factors. As a result, our approach successfully identified biologically relevant connections that were not previously known to the predictive models. Additionally, we applied a genome-wide association study (GWAS) on key metabolomics features, revealing a link between vascular dementia and smoking. Although being a well-established epidemiological relationship, this link was not embedded in the model's training data, which validated the method's ability to extract meaningful signals. Furthermore, by integrating survival models as inputs in the 10K data, we uncovered associations between metabolic substances and obesity, demonstrating the ability to infer disease risk for future patients without requiring direct outcome labels. These findings highlight the potential of leveraging external bio-banks to extract valuable biomedical insights, even in data-limited scenarios. Our results demonstrate that machine learning models trained on smaller datasets can still be used to uncover real biological associations when carefully integrated with survival analysis and genetic studies.
SGAC: A Graph Neural Network Framework for Imbalanced and Structure-Aware AMP Classification
Dec 20, 2024Abstract:Classifying antimicrobial peptides(AMPs) from the vast array of peptides mined from metagenomic sequencing data is a significant approach to addressing the issue of antibiotic resistance. However, current AMP classification methods, primarily relying on sequence-based data, neglect the spatial structure of peptides, thereby limiting the accurate classification of AMPs. Additionally, the number of known AMPs is significantly lower than that of non-AMPs, leading to imbalanced datasets that reduce predictive accuracy for AMPs. To alleviate these two limitations, we first employ Omegafold to predict the three-dimensional spatial structures of AMPs and non-AMPs, constructing peptide graphs based on the amino acids' C$_\alpha$ positions. Building upon this, we propose a novel classification model named Spatial GNN-based AMP Classifier (SGAC). Our SGAC model employs a graph encoder based on Graph Neural Networks (GNNs) to process peptide graphs, generating high-dimensional representations that capture essential features from the three-dimensional spatial structure of amino acids. Then, to address the inherent imbalanced datasets, SGAC first incorporates Weight-enhanced Contrastive Learning, which clusters similar peptides while ensuring separation between dissimilar ones, using weighted contributions to emphasize AMP-specific features. Furthermore, SGAC employs Weight-enhanced Pseudo-label Distillation to dynamically generate high-confidence pseudo labels for ambiguous peptides, further refining predictions and promoting balanced learning between AMPs and non-AMPs. Experiments on publicly available AMP and non-AMP datasets demonstrate that SGAC significantly outperforms traditional sequence-based methods and achieves state-of-the-art performance among graph-based models, validating its effectiveness in AMP classification.
Toward AI-Driven Digital Organism: Multiscale Foundation Models for Predicting, Simulating and Programming Biology at All Levels
Dec 09, 2024



Abstract:We present an approach of using AI to model and simulate biology and life. Why is it important? Because at the core of medicine, pharmacy, public health, longevity, agriculture and food security, environmental protection, and clean energy, it is biology at work. Biology in the physical world is too complex to manipulate and always expensive and risky to tamper with. In this perspective, we layout an engineering viable approach to address this challenge by constructing an AI-Driven Digital Organism (AIDO), a system of integrated multiscale foundation models, in a modular, connectable, and holistic fashion to reflect biological scales, connectedness, and complexities. An AIDO opens up a safe, affordable and high-throughput alternative platform for predicting, simulating and programming biology at all levels from molecules to cells to individuals. We envision that an AIDO is poised to trigger a new wave of better-guided wet-lab experimentation and better-informed first-principle reasoning, which can eventually help us better decode and improve life.
Causal Representation Learning from Multimodal Biological Observations
Nov 10, 2024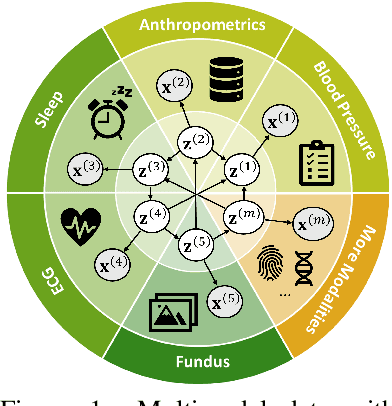

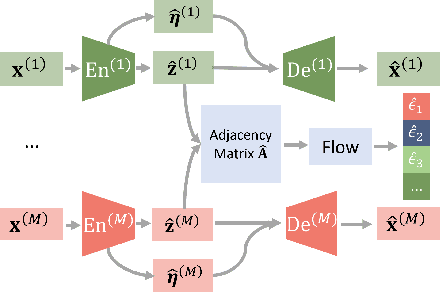
Abstract:Prevalent in biological applications (e.g., human phenotype measurements), multimodal datasets can provide valuable insights into the underlying biological mechanisms. However, current machine learning models designed to analyze such datasets still lack interpretability and theoretical guarantees, which are essential to biological applications. Recent advances in causal representation learning have shown promise in uncovering the interpretable latent causal variables with formal theoretical certificates. Unfortunately, existing works for multimodal distributions either rely on restrictive parametric assumptions or provide rather coarse identification results, limiting their applicability to biological research which favors a detailed understanding of the mechanisms. In this work, we aim to develop flexible identification conditions for multimodal data and principled methods to facilitate the understanding of biological datasets. Theoretically, we consider a flexible nonparametric latent distribution (c.f., parametric assumptions in prior work) permitting causal relationships across potentially different modalities. We establish identifiability guarantees for each latent component, extending the subspace identification results from prior work. Our key theoretical ingredient is the structural sparsity of the causal connections among distinct modalities, which, as we will discuss, is natural for a large collection of biological systems. Empirically, we propose a practical framework to instantiate our theoretical insights. We demonstrate the effectiveness of our approach through extensive experiments on both numerical and synthetic datasets. Results on a real-world human phenotype dataset are consistent with established medical research, validating our theoretical and methodological framework.
Generative AI Enables Medical Image Segmentation in Ultra Low-Data Regimes
Aug 30, 2024Abstract:Semantic segmentation of medical images is pivotal in applications like disease diagnosis and treatment planning. While deep learning has excelled in automating this task, a major hurdle is the need for numerous annotated segmentation masks, which are resource-intensive to produce due to the required expertise and time. This scenario often leads to ultra low-data regimes, where annotated images are extremely limited, posing significant challenges for the generalization of conventional deep learning methods on test images. To address this, we introduce a generative deep learning framework, which uniquely generates high-quality paired segmentation masks and medical images, serving as auxiliary data for training robust models in data-scarce environments. Unlike traditional generative models that treat data generation and segmentation model training as separate processes, our method employs multi-level optimization for end-to-end data generation. This approach allows segmentation performance to directly influence the data generation process, ensuring that the generated data is specifically tailored to enhance the performance of the segmentation model. Our method demonstrated strong generalization performance across 9 diverse medical image segmentation tasks and on 16 datasets, in ultra-low data regimes, spanning various diseases, organs, and imaging modalities. When applied to various segmentation models, it achieved performance improvements of 10-20\% (absolute), in both same-domain and out-of-domain scenarios. Notably, it requires 8 to 20 times less training data than existing methods to achieve comparable results. This advancement significantly improves the feasibility and cost-effectiveness of applying deep learning in medical imaging, particularly in scenarios with limited data availability.
From Glucose Patterns to Health Outcomes: A Generalizable Foundation Model for Continuous Glucose Monitor Data Analysis
Aug 20, 2024Abstract:Recent advances in self-supervised learning enabled novel medical AI models, known as foundation models (FMs) that offer great potential for characterizing health from diverse biomedical data. Continuous glucose monitoring (CGM) provides rich, temporal data on glycemic patterns, but its full potential for predicting broader health outcomes remains underutilized. Here, we present GluFormer, a generative foundation model on biomedical temporal data based on a transformer architecture, and trained on over 10 million CGM measurements from 10,812 non-diabetic individuals. We tokenized the CGM training data and trained GluFormer using next token prediction in a generative, autoregressive manner. We demonstrate that GluFormer generalizes effectively to 15 different external datasets, including 4936 individuals across 5 different geographical regions, 6 different CGM devices, and several metabolic disorders, including normoglycemic, prediabetic, and diabetic populations, as well as those with gestational diabetes and obesity. GluFormer produces embeddings which outperform traditional CGM analysis tools, and achieves high Pearson correlations in predicting clinical parameters such as HbA1c, liver-related parameters, blood lipids, and sleep-related indices. Notably, GluFormer can also predict onset of future health outcomes even 4 years in advance. We also show that CGM embeddings from pre-intervention periods in Randomized Clinical Trials (RCTs) outperform other methods in predicting primary and secondary outcomes. When integrating dietary data into GluFormer, we show that the enhanced model can accurately generate CGM data based only on dietary intake data, simulate outcomes of dietary interventions, and predict individual responses to specific foods. Overall, we show that GluFormer accurately predicts health outcomes which generalize across different populations metabolic conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge