Daphne Koller
Stanford University
Peptide-Spectra Matching from Weak Supervision
Aug 22, 2018



Abstract:As in many other scientific domains, we face a fundamental problem when using machine learning to identify proteins from mass spectrometry data: large ground truth datasets mapping inputs to correct outputs are extremely difficult to obtain. Instead, we have access to imperfect hand-coded models crafted by domain experts. In this paper, we apply deep neural networks to an important step of the protein identification problem, the pairing of mass spectra with short sequences of amino acids called peptides. We train our model to differentiate between top scoring results from a state-of-the art classical system and hard-negative second and third place results. Our resulting model is much better at identifying peptides with spectra than the model used to generate its training data. In particular, we achieve a 43% improvement over standard matching methods and a 10% improvement over a combination of the matching method and an industry standard cross-spectra reranking tool. Importantly, in a more difficult experimental regime that reflects current challenges facing biologists, our advantage over the previous state-of-the-art grows to 15% even after reranking. We believe this approach will generalize to other challenging scientific problems.
Inferring Multi-Dimensional Rates of Aging from Cross-Sectional Data
Jul 12, 2018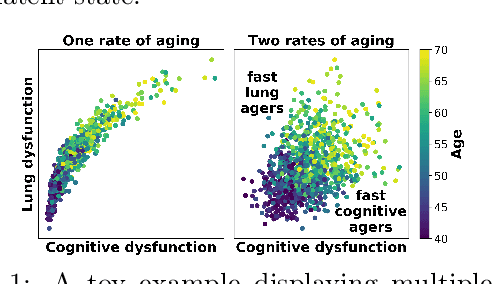
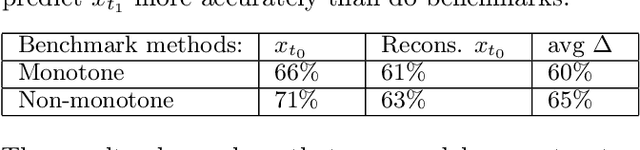
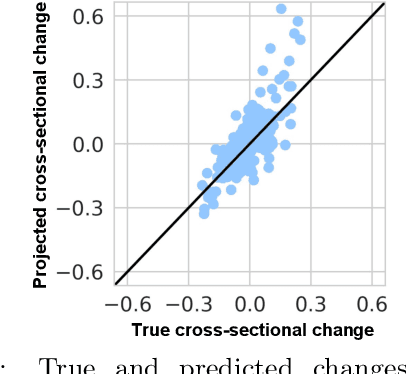
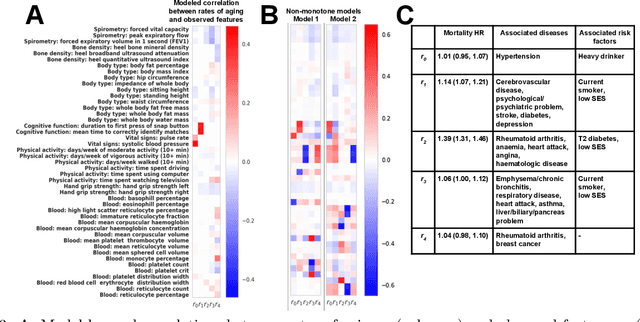
Abstract:Modeling how individuals evolve over time is a fundamental problem in the natural and social sciences. However, existing datasets are often cross-sectional with each individual only observed at a single timepoint, making inference of temporal dynamics hard. Motivated by the study of human aging, we present a model that can learn temporal dynamics from cross-sectional data. Our model represents each individual with a low-dimensional latent state that consists of 1) a dynamic vector $rt$ that evolves linearly with time $t$, where $r$ is an individual-specific "rate of aging" vector, and 2) a static vector $b$ that captures time-independent variation. Observed features are a non-linear function of $rt$ and $b$. We prove that constraining the mapping between $rt$ and a subset of the observed features to be order-isomorphic yields a model class that is identifiable if the distribution of time-independent variation is known. Our model correctly recovers the latent rate vector $r$ in realistic synthetic data. Applied to the UK Biobank human health dataset, our model accurately reconstructs the observed data while learning interpretable rates of aging $r$ that are positively associated with diseases, mortality, and aging risk factors.
Proceedings of the Seventeenth Conference on Uncertainty in Artificial Intelligence (2001)
Aug 28, 2014Abstract:This is the Proceedings of the Seventeenth Conference on Uncertainty in Artificial Intelligence, which was held in Seattle, WA, August 2-5 2001
Tuned Models of Peer Assessment in MOOCs
Jul 09, 2013



Abstract:In massive open online courses (MOOCs), peer grading serves as a critical tool for scaling the grading of complex, open-ended assignments to courses with tens or hundreds of thousands of students. But despite promising initial trials, it does not always deliver accurate results compared to human experts. In this paper, we develop algorithms for estimating and correcting for grader biases and reliabilities, showing significant improvement in peer grading accuracy on real data with 63,199 peer grades from Coursera's HCI course offerings --- the largest peer grading networks analysed to date. We relate grader biases and reliabilities to other student factors such as student engagement, performance as well as commenting style. We also show that our model can lead to more intelligent assignment of graders to gradees.
Probability Estimation in Face of Irrelevant Information
Mar 20, 2013Abstract:In this paper, we consider one aspect of the problem of applying decision theory to the design of agents that learn how to make decisions under uncertainty. This aspect concerns how an agent can estimate probabilities for the possible states of the world, given that it only makes limited observations before committing to a decision. We show that the naive application of statistical tools can be improved upon if the agent can determine which of his observations are truly relevant to the estimation problem at hand. We give a framework in which such determinations can be made, and define an estimation procedure to use them. Our framework also suggests several extensions, which show how additional knowledge can be used to improve tile estimation procedure still further.
Generating New Beliefs From Old
Feb 27, 2013Abstract:In previous work [BGHK92, BGHK93], we have studied the random-worlds approach -- a particular (and quite powerful) method for generating degrees of belief (i.e., subjective probabilities) from a knowledge base consisting of objective (first-order, statistical, and default) information. But allowing a knowledge base to contain only objective information is sometimes limiting. We occasionally wish to include information about degrees of belief in the knowledge base as well, because there are contexts in which old beliefs represent important information that should influence new beliefs. In this paper, we describe three quite general techniques for extending a method that generates degrees of belief from objective information to one that can make use of degrees of belief as well. All of our techniques are bloused on well-known approaches, such as cross-entropy. We discuss general connections between the techniques and in particular show that, although conceptually and technically quite different, all of the techniques give the same answer when applied to the random-worlds method.
Stochastic Simulation Algorithms for Dynamic Probabilistic Networks
Feb 20, 2013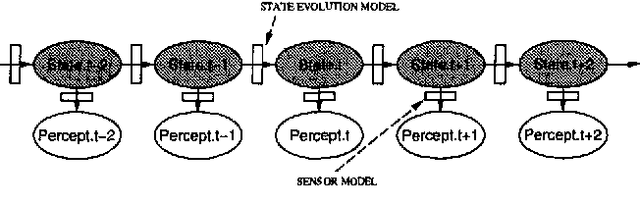
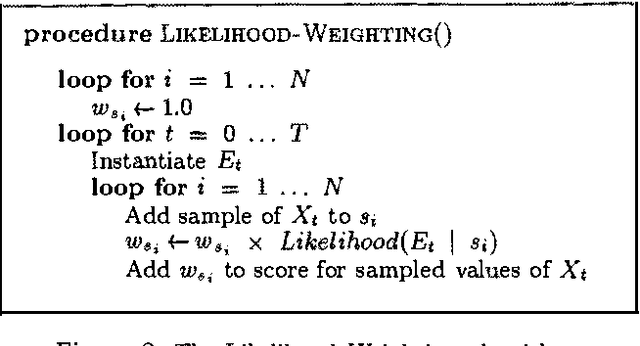
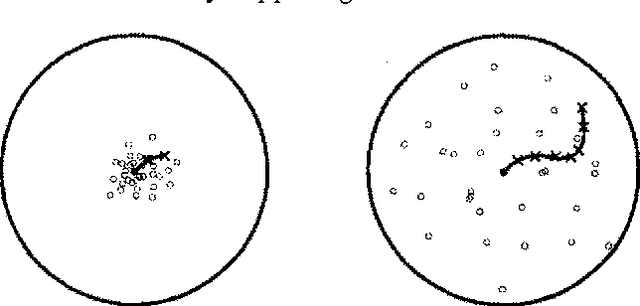
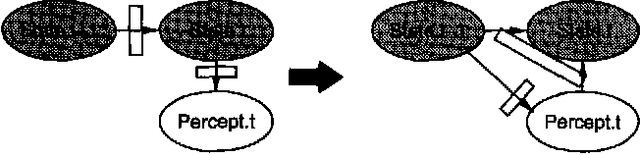
Abstract:Stochastic simulation algorithms such as likelihood weighting often give fast, accurate approximations to posterior probabilities in probabilistic networks, and are the methods of choice for very large networks. Unfortunately, the special characteristics of dynamic probabilistic networks (DPNs), which are used to represent stochastic temporal processes, mean that standard simulation algorithms perform very poorly. In essence, the simulation trials diverge further and further from reality as the process is observed over time. In this paper, we present simulation algorithms that use the evidence observed at each time step to push the set of trials back towards reality. The first algorithm, "evidence reversal" (ER) restructures each time slice of the DPN so that the evidence nodes for the slice become ancestors of the state variables. The second algorithm, called "survival of the fittest" sampling (SOF), "repopulates" the set of trials at each time step using a stochastic reproduction rate weighted by the likelihood of the evidence according to each trial. We compare the performance of each algorithm with likelihood weighting on the original network, and also investigate the benefits of combining the ER and SOF methods. The ER/SOF combination appears to maintain bounded error independent of the number of time steps in the simulation.
Context-Specific Independence in Bayesian Networks
Feb 13, 2013



Abstract:Bayesian networks provide a language for qualitatively representing the conditional independence properties of a distribution. This allows a natural and compact representation of the distribution, eases knowledge acquisition, and supports effective inference algorithms. It is well-known, however, that there are certain independencies that we cannot capture qualitatively within the Bayesian network structure: independencies that hold only in certain contexts, i.e., given a specific assignment of values to certain variables. In this paper, we propose a formal notion of context-specific independence (CSI), based on regularities in the conditional probability tables (CPTs) at a node. We present a technique, analogous to (and based on) d-separation, for determining when such independence holds in a given network. We then focus on a particular qualitative representation scheme - tree-structured CPTs - for capturing CSI. We suggest ways in which this representation can be used to support effective inference algorithms. In particular, we present a structural decomposition of the resulting network which can improve the performance of clustering algorithms, and an alternative algorithm based on cutset conditioning.
Nonuniform Dynamic Discretization in Hybrid Networks
Feb 06, 2013



Abstract:We consider probabilistic inference in general hybrid networks, which include continuous and discrete variables in an arbitrary topology. We reexamine the question of variable discretization in a hybrid network aiming at minimizing the information loss induced by the discretization. We show that a nonuniform partition across all variables as opposed to uniform partition of each variable separately reduces the size of the data structures needed to represent a continuous function. We also provide a simple but efficient procedure for nonuniform partition. To represent a nonuniform discretization in the computer memory, we introduce a new data structure, which we call a Binary Split Partition (BSP) tree. We show that BSP trees can be an exponential factor smaller than the data structures in the standard uniform discretization in multiple dimensions and show how the BSP trees can be used in the standard join tree algorithm. We show that the accuracy of the inference process can be significantly improved by adjusting discretization with evidence. We construct an iterative anytime algorithm that gradually improves the quality of the discretization and the accuracy of the answer on a query. We provide empirical evidence that the algorithm converges.
Object-Oriented Bayesian Networks
Feb 06, 2013

Abstract:Bayesian networks provide a modeling language and associated inference algorithm for stochastic domains. They have been successfully applied in a variety of medium-scale applications. However, when faced with a large complex domain, the task of modeling using Bayesian networks begins to resemble the task of programming using logical circuits. In this paper, we describe an object-oriented Bayesian network (OOBN) language, which allows complex domains to be described in terms of inter-related objects. We use a Bayesian network fragment to describe the probabilistic relations between the attributes of an object. These attributes can themselves be objects, providing a natural framework for encoding part-of hierarchies. Classes are used to provide a reusable probabilistic model which can be applied to multiple similar objects. Classes also support inheritance of model fragments from a class to a subclass, allowing the common aspects of related classes to be defined only once. Our language has clear declarative semantics: an OOBN can be interpreted as a stochastic functional program, so that it uniquely specifies a probabilistic model. We provide an inference algorithm for OOBNs, and show that much of the structural information encoded by an OOBN--particularly the encapsulation of variables within an object and the reuse of model fragments in different contexts--can also be used to speed up the inference process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge