David Wilson
Generative AI Enables Medical Image Segmentation in Ultra Low-Data Regimes
Aug 30, 2024Abstract:Semantic segmentation of medical images is pivotal in applications like disease diagnosis and treatment planning. While deep learning has excelled in automating this task, a major hurdle is the need for numerous annotated segmentation masks, which are resource-intensive to produce due to the required expertise and time. This scenario often leads to ultra low-data regimes, where annotated images are extremely limited, posing significant challenges for the generalization of conventional deep learning methods on test images. To address this, we introduce a generative deep learning framework, which uniquely generates high-quality paired segmentation masks and medical images, serving as auxiliary data for training robust models in data-scarce environments. Unlike traditional generative models that treat data generation and segmentation model training as separate processes, our method employs multi-level optimization for end-to-end data generation. This approach allows segmentation performance to directly influence the data generation process, ensuring that the generated data is specifically tailored to enhance the performance of the segmentation model. Our method demonstrated strong generalization performance across 9 diverse medical image segmentation tasks and on 16 datasets, in ultra-low data regimes, spanning various diseases, organs, and imaging modalities. When applied to various segmentation models, it achieved performance improvements of 10-20\% (absolute), in both same-domain and out-of-domain scenarios. Notably, it requires 8 to 20 times less training data than existing methods to achieve comparable results. This advancement significantly improves the feasibility and cost-effectiveness of applying deep learning in medical imaging, particularly in scenarios with limited data availability.
Extended Agriculture-Vision: An Extension of a Large Aerial Image Dataset for Agricultural Pattern Analysis
Mar 04, 2023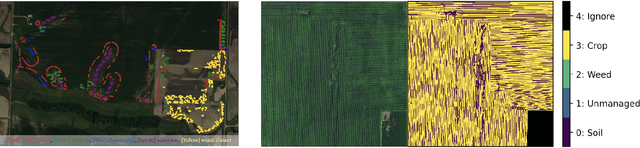
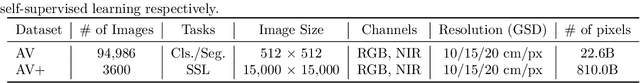
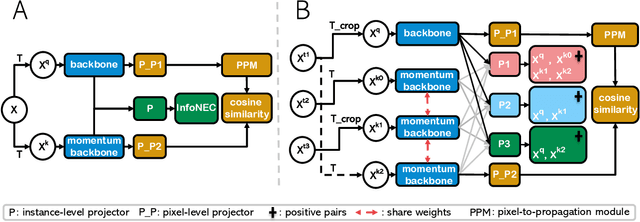
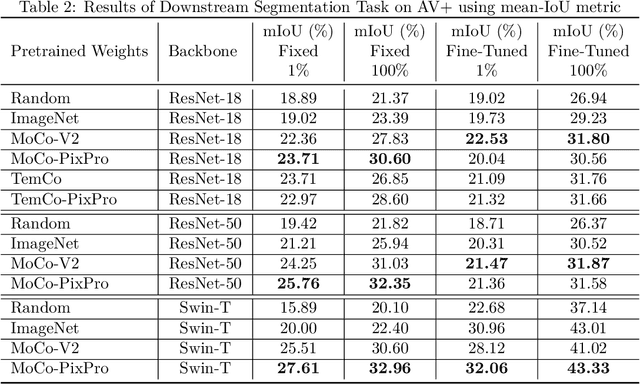
Abstract:A key challenge for much of the machine learning work on remote sensing and earth observation data is the difficulty in acquiring large amounts of accurately labeled data. This is particularly true for semantic segmentation tasks, which are much less common in the remote sensing domain because of the incredible difficulty in collecting precise, accurate, pixel-level annotations at scale. Recent efforts have addressed these challenges both through the creation of supervised datasets as well as the application of self-supervised methods. We continue these efforts on both fronts. First, we generate and release an improved version of the Agriculture-Vision dataset (Chiu et al., 2020b) to include raw, full-field imagery for greater experimental flexibility. Second, we extend this dataset with the release of 3600 large, high-resolution (10cm/pixel), full-field, red-green-blue and near-infrared images for pre-training. Third, we incorporate the Pixel-to-Propagation Module Xie et al. (2021b) originally built on the SimCLR framework into the framework of MoCo-V2 Chen et al.(2020b). Finally, we demonstrate the usefulness of this data by benchmarking different contrastive learning approaches on both downstream classification and semantic segmentation tasks. We explore both CNN and Swin Transformer Liu et al. (2021a) architectures within different frameworks based on MoCo-V2. Together, these approaches enable us to better detect key agricultural patterns of interest across a field from aerial imagery so that farmers may be alerted to problematic areas in a timely fashion to inform their management decisions. Furthermore, the release of these datasets will support numerous avenues of research for computer vision in remote sensing for agriculture.
* Dataset: https://github.com/jingwu6/Extended-Agriculture-Vision-Dataset Video: https://youtu.be/2xaKxUpY4iQ
Superpixels and Graph Convolutional Neural Networks for Efficient Detection of Nutrient Deficiency Stress from Aerial Imagery
Apr 22, 2021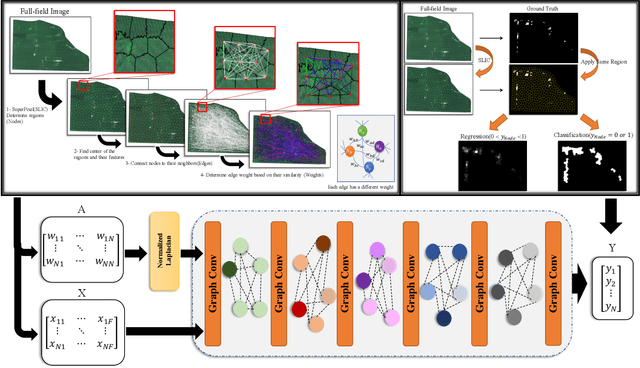
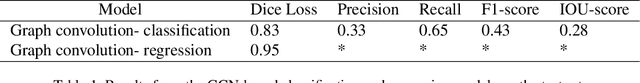
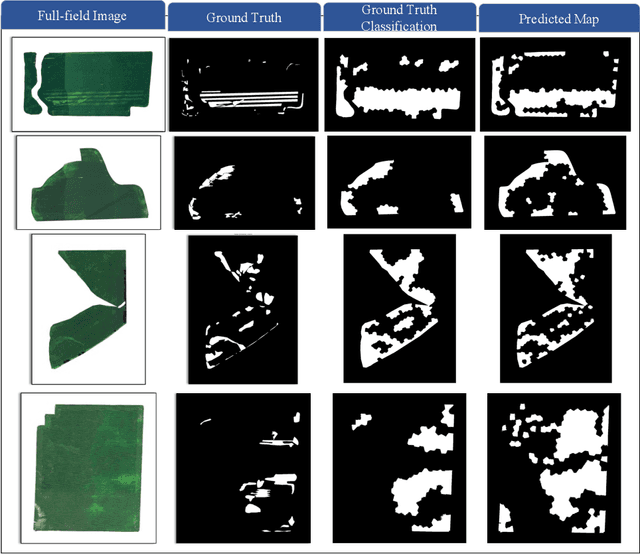
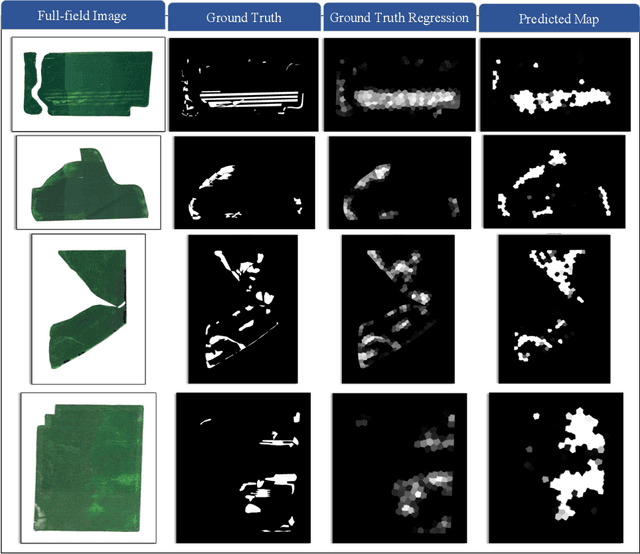
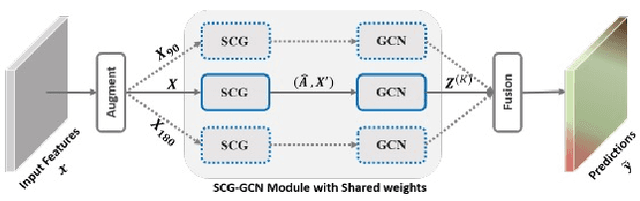
Abstract:Advances in remote sensing technology have led to the capture of massive amounts of data. Increased image resolution, more frequent revisit times, and additional spectral channels have created an explosion in the amount of data that is available to provide analyses and intelligence across domains, including agriculture. However, the processing of this data comes with a cost in terms of computation time and money, both of which must be considered when the goal of an algorithm is to provide real-time intelligence to improve efficiencies. Specifically, we seek to identify nutrient deficient areas from remotely sensed data to alert farmers to regions that require attention; detection of nutrient deficient areas is a key task in precision agriculture as farmers must quickly respond to struggling areas to protect their harvests. Past methods have focused on pixel-level classification (i.e. semantic segmentation) of the field to achieve these tasks, often using deep learning models with tens-of-millions of parameters. In contrast, we propose a much lighter graph-based method to perform node-based classification. We first use Simple Linear Iterative Cluster (SLIC) to produce superpixels across the field. Then, to perform segmentation across the non-Euclidean domain of superpixels, we leverage a Graph Convolutional Neural Network (GCN). This model has 4-orders-of-magnitude fewer parameters than a CNN model and trains in a matter of minutes.
The 1st Agriculture-Vision Challenge: Methods and Results
Apr 23, 2020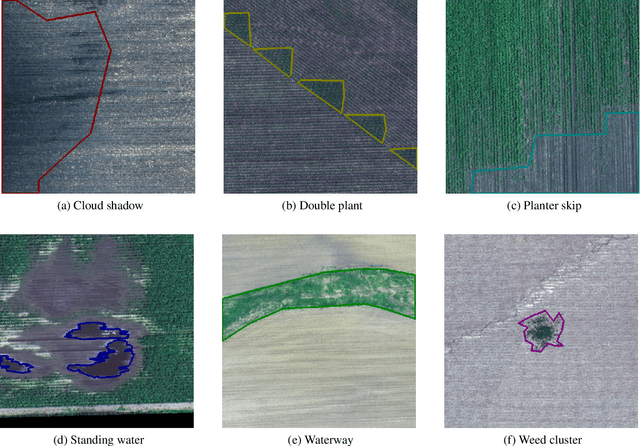
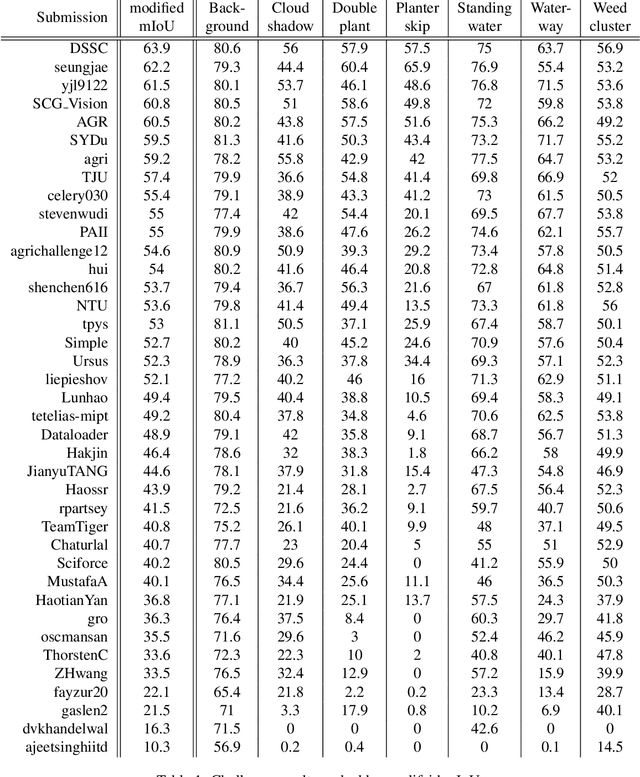
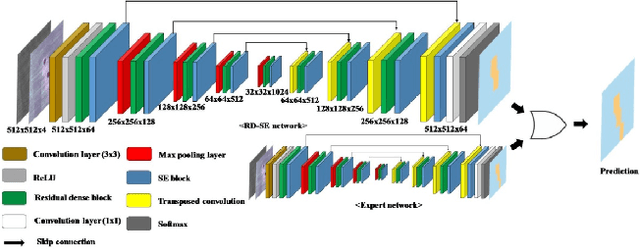
Abstract:The first Agriculture-Vision Challenge aims to encourage research in developing novel and effective algorithms for agricultural pattern recognition from aerial images, especially for the semantic segmentation task associated with our challenge dataset. Around 57 participating teams from various countries compete to achieve state-of-the-art in aerial agriculture semantic segmentation. The Agriculture-Vision Challenge Dataset was employed, which comprises of 21,061 aerial and multi-spectral farmland images. This paper provides a summary of notable methods and results in the challenge. Our submission server and leaderboard will continue to open for researchers that are interested in this challenge dataset and task; the link can be found here.
Agriculture-Vision: A Large Aerial Image Database for Agricultural Pattern Analysis
Jan 05, 2020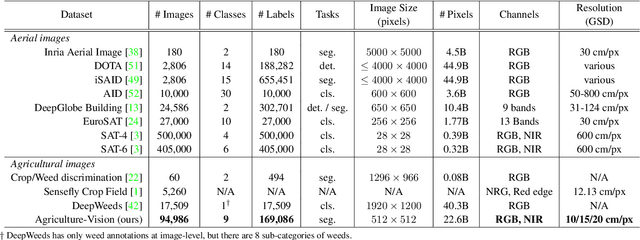


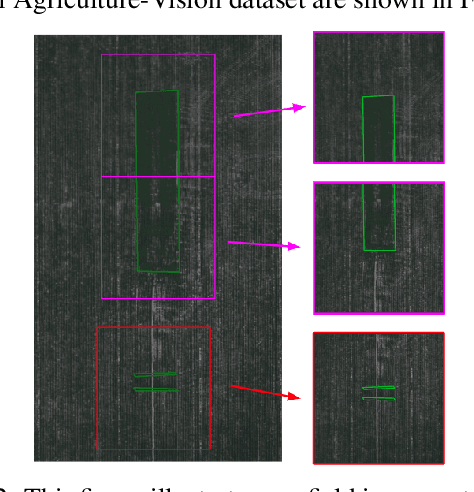
Abstract:The success of deep learning in visual recognition tasks has driven advancements in multiple fields of research. Particularly, increasing attention has been drawn towards its application in agriculture. Nevertheless, while visual pattern recognition on farmlands carries enormous economic values, little progress has been made to merge computer vision and crop sciences due to the lack of suitable agricultural image datasets. Meanwhile, problems in agriculture also pose new challenges in computer vision. For example, semantic segmentation of aerial farmland images requires inference over extremely large-size images with extreme annotation sparsity. These challenges are not present in most of the common object datasets, and we show that they are more challenging than many other aerial image datasets. To encourage research in computer vision for agriculture, we present Agriculture-Vision: a large-scale aerial farmland image dataset for semantic segmentation of agricultural patterns. We collected 94,986 high-quality aerial images from 3,432 farmlands across the US, where each image consists of RGB and Near-infrared (NIR) channels with resolution as high as 10 cm per pixel. We annotate nine types of field anomaly patterns that are most important to farmers. As a pilot study of aerial agricultural semantic segmentation, we perform comprehensive experiments using popular semantic segmentation models; we also propose an effective model designed for aerial agricultural pattern recognition. Our experiments demonstrate several challenges Agriculture-Vision poses to both the computer vision and agriculture communities. Future versions of this dataset will include even more aerial images, anomaly patterns and image channels. More information at https://www.agriculture-vision.com.
Generation of Virtual Dual Energy Images from Standard Single-Shot Radiographs using Multi-scale and Conditional Adversarial Network
Oct 22, 2018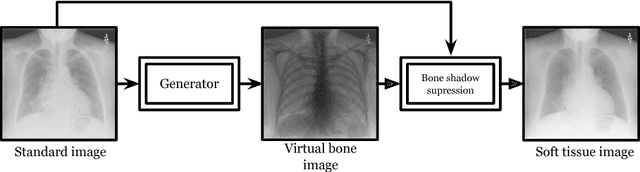
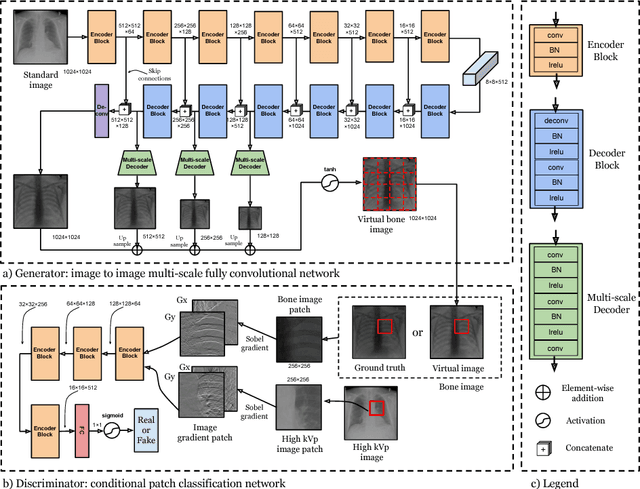
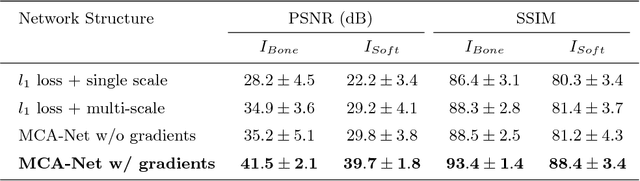
Abstract:Dual-energy (DE) chest radiographs provide greater diagnostic information than standard radiographs by separating the image into bone and soft tissue, revealing suspicious lesions which may otherwise be obstructed from view. However, acquisition of DE images requires two physical scans, necessitating specialized hardware and processing, and images are prone to motion artifact. Generation of virtual DE images from standard, single-shot chest radiographs would expand the diagnostic value of standard radiographs without changing the acquisition procedure. We present a Multi-scale Conditional Adversarial Network (MCA-Net) which produces high-resolution virtual DE bone images from standard, single-shot chest radiographs. Our proposed MCA-Net is trained using the adversarial network so that it learns sharp details for the production of high-quality bone images. Then, the virtual DE soft tissue image is generated by processing the standard radiograph with the virtual bone image using a cross projection transformation. Experimental results from 210 patient DE chest radiographs demonstrated that the algorithm can produce high-quality virtual DE chest radiographs. Important structures were preserved, such as coronary calcium in bone images and lung lesions in soft tissue images. The average structure similarity index and the peak signal to noise ratio of the produced bone images in testing data were 96.4 and 41.5, which are significantly better than results from previous methods. Furthermore, our clinical evaluation results performed on the publicly available dataset indicates the clinical values of our algorithms. Thus, our algorithm can produce high-quality DE images that are potentially useful for radiologists, computer-aided diagnostics, and other diagnostic tasks.
Using Machine Learning to Improve Cylindrical Algebraic Decomposition
Apr 26, 2018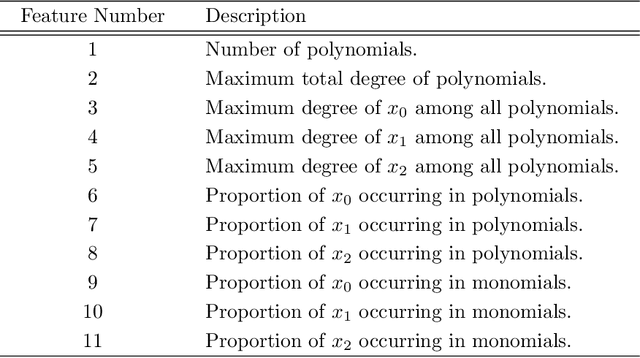
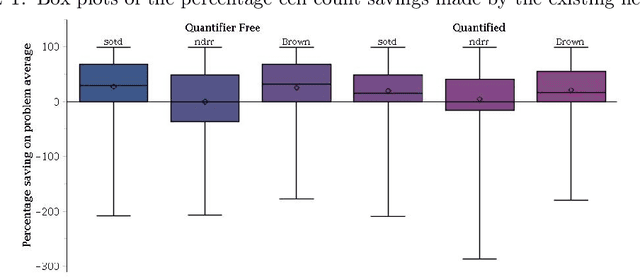
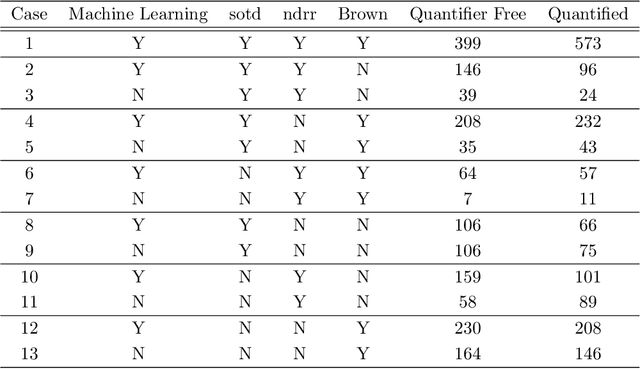
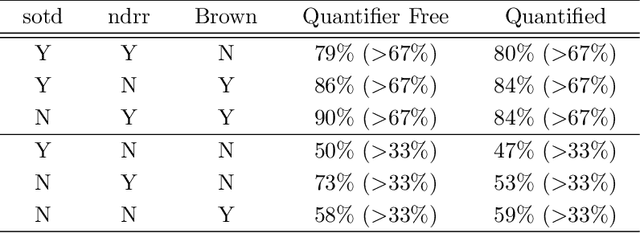
Abstract:Cylindrical Algebraic Decomposition (CAD) is a key tool in computational algebraic geometry, best known as a procedure to enable Quantifier Elimination over real-closed fields. However, it has a worst case complexity doubly exponential in the size of the input, which is often encountered in practice. It has been observed that for many problems a change in algorithm settings or problem formulation can cause huge differences in runtime costs, changing problem instances from intractable to easy. A number of heuristics have been developed to help with such choices, but the complicated nature of the geometric relationships involved means these are imperfect and can sometimes make poor choices. We investigate the use of machine learning (specifically support vector machines) to make such choices instead. Machine learning is the process of fitting a computer model to a complex function based on properties learned from measured data. In this paper we apply it in two case studies: the first to select between heuristics for choosing a CAD variable ordering; the second to identify when a CAD problem instance would benefit from Groebner Basis preconditioning. These appear to be the first such applications of machine learning to Symbolic Computation. We demonstrate in both cases that the machine learned choice outperforms human developed heuristics.
Applying machine learning to the problem of choosing a heuristic to select the variable ordering for cylindrical algebraic decomposition
Apr 25, 2014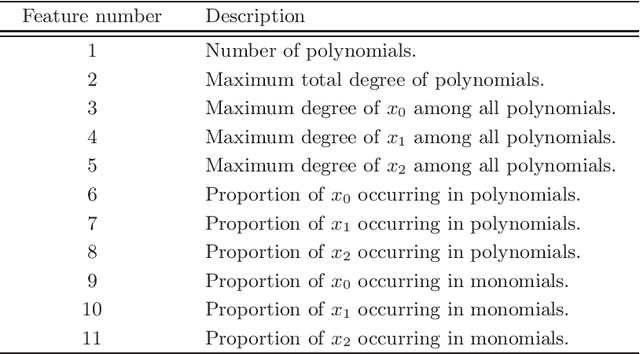
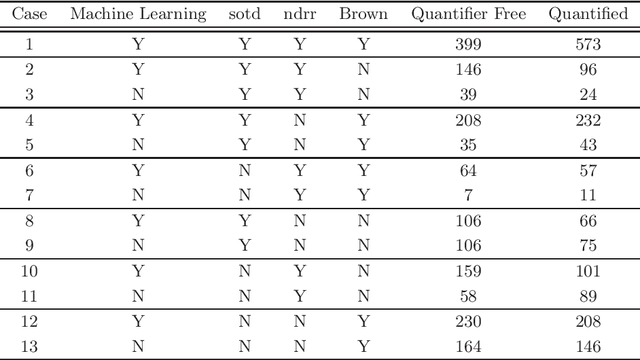
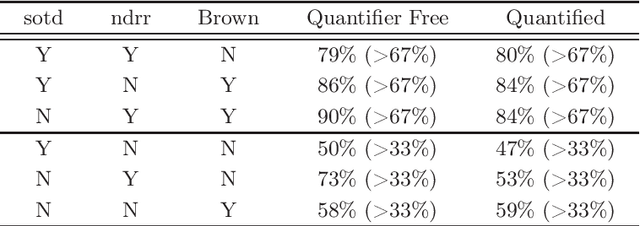

Abstract:Cylindrical algebraic decomposition(CAD) is a key tool in computational algebraic geometry, particularly for quantifier elimination over real-closed fields. When using CAD, there is often a choice for the ordering placed on the variables. This can be important, with some problems infeasible with one variable ordering but easy with another. Machine learning is the process of fitting a computer model to a complex function based on properties learned from measured data. In this paper we use machine learning (specifically a support vector machine) to select between heuristics for choosing a variable ordering, outperforming each of the separate heuristics.
* 16 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge