Padchest
Papers and Code
Bi-MCQ: Reformulating Vision-Language Alignment for Negation Understanding
Jan 30, 2026Recent vision-language models (VLMs) achieve strong zero-shot performance via large-scale image-text pretraining and have been widely adopted in medical image analysis. However, existing VLMs remain notably weak at understanding negated clinical statements, largely due to contrastive alignment objectives that treat negation as a minor linguistic variation rather than a meaning-inverting operator. In multi-label settings, prompt-based InfoNCE fine-tuning further reinforces easy-positive image-prompt alignments, limiting effective learning of disease absence. To overcome these limitations, we reformulate vision-language alignment as a conditional semantic comparison problem, which is instantiated through a bi-directional multiple-choice learning framework(Bi-MCQ). By jointly training Image-to-Text and Text-to-Image MCQ tasks with affirmative, negative, and mixed prompts, our method implements fine-tuning as conditional semantic comparison instead of global similarity maximization. We further introduce direction-specific Cross-Attention fusion modules to address asymmetric cues required by bi-directional reasoning and reduce alignment interference. Experiments on ChestXray14, Open-I, CheXpert, and PadChest show that Bi-MCQ improves negation understanding by up to 0.47 AUC over the zero-shot performance of the state-of-the-art CARZero model, while achieving up to a 0.08 absolute gain on positive-negative combined (PNC) evaluation. Additionally, Bi-MCQ reduces the affirmative-negative AUC gap by an average of 0.12 compared to InfoNCE-based fine-tuning, demonstrating that objective reformulation can substantially enhance negation understanding in medical VLMs.
Limitations of Public Chest Radiography Datasets for Artificial Intelligence: Label Quality, Domain Shift, Bias and Evaluation Challenges
Sep 18, 2025



Artificial intelligence has shown significant promise in chest radiography, where deep learning models can approach radiologist-level diagnostic performance. Progress has been accelerated by large public datasets such as MIMIC-CXR, ChestX-ray14, PadChest, and CheXpert, which provide hundreds of thousands of labelled images with pathology annotations. However, these datasets also present important limitations. Automated label extraction from radiology reports introduces errors, particularly in handling uncertainty and negation, and radiologist review frequently disagrees with assigned labels. In addition, domain shift and population bias restrict model generalisability, while evaluation practices often overlook clinically meaningful measures. We conduct a systematic analysis of these challenges, focusing on label quality, dataset bias, and domain shift. Our cross-dataset domain shift evaluation across multiple model architectures revealed substantial external performance degradation, with pronounced reductions in AUPRC and F1 scores relative to internal testing. To assess dataset bias, we trained a source-classification model that distinguished datasets with near-perfect accuracy, and performed subgroup analyses showing reduced performance for minority age and sex groups. Finally, expert review by two board-certified radiologists identified significant disagreement with public dataset labels. Our findings highlight important clinical weaknesses of current benchmarks and emphasise the need for clinician-validated datasets and fairer evaluation frameworks.
Understanding Dataset Bias in Medical Imaging: A Case Study on Chest X-rays
Jul 10, 2025Recent work has revisited the infamous task Name that dataset and established that in non-medical datasets, there is an underlying bias and achieved high Accuracies on the dataset origin task. In this work, we revisit the same task applied to popular open-source chest X-ray datasets. Medical images are naturally more difficult to release for open-source due to their sensitive nature, which has led to certain open-source datasets being extremely popular for research purposes. By performing the same task, we wish to explore whether dataset bias also exists in these datasets. % We deliberately try to increase the difficulty of the task by dataset transformations. We apply simple transformations of the datasets to try to identify bias. Given the importance of AI applications in medical imaging, it's vital to establish whether modern methods are taking shortcuts or are focused on the relevant pathology. We implement a range of different network architectures on the datasets: NIH, CheXpert, MIMIC-CXR and PadChest. We hope this work will encourage more explainable research being performed in medical imaging and the creation of more open-source datasets in the medical domain. The corresponding code will be released upon acceptance.
Mask of truth: model sensitivity to unexpected regions of medical images
Dec 05, 2024The development of larger models for medical image analysis has led to increased performance. However, it also affected our ability to explain and validate model decisions. Models can use non-relevant parts of images, also called spurious correlations or shortcuts, to obtain high performance on benchmark datasets but fail in real-world scenarios. In this work, we challenge the capacity of convolutional neural networks (CNN) to classify chest X-rays and eye fundus images while masking out clinically relevant parts of the image. We show that all models trained on the PadChest dataset, irrespective of the masking strategy, are able to obtain an Area Under the Curve (AUC) above random. Moreover, the models trained on full images obtain good performance on images without the region of interest (ROI), even superior to the one obtained on images only containing the ROI. We also reveal a possible spurious correlation in the Chaksu dataset while the performances are more aligned with the expectation of an unbiased model. We go beyond the performance analysis with the usage of the explainability method SHAP and the analysis of embeddings. We asked a radiology resident to interpret chest X-rays under different masking to complement our findings with clinical knowledge. Our code is available at https://github.com/TheoSourget/MMC_Masking and https://github.com/TheoSourget/MMC_Masking_EyeFundus
PadChest-GR: A Bilingual Chest X-ray Dataset for Grounded Radiology Report Generation
Nov 07, 2024



Radiology report generation (RRG) aims to create free-text radiology reports from clinical imaging. Grounded radiology report generation (GRRG) extends RRG by including the localisation of individual findings on the image. Currently, there are no manually annotated chest X-ray (CXR) datasets to train GRRG models. In this work, we present a dataset called PadChest-GR (Grounded-Reporting) derived from PadChest aimed at training GRRG models for CXR images. We curate a public bi-lingual dataset of 4,555 CXR studies with grounded reports (3,099 abnormal and 1,456 normal), each containing complete lists of sentences describing individual present (positive) and absent (negative) findings in English and Spanish. In total, PadChest-GR contains 7,037 positive and 3,422 negative finding sentences. Every positive finding sentence is associated with up to two independent sets of bounding boxes labelled by different readers and has categorical labels for finding type, locations, and progression. To the best of our knowledge, PadChest-GR is the first manually curated dataset designed to train GRRG models for understanding and interpreting radiological images and generated text. By including detailed localization and comprehensive annotations of all clinically relevant findings, it provides a valuable resource for developing and evaluating GRRG models from CXR images. PadChest-GR can be downloaded under request from https://bimcv.cipf.es/bimcv-projects/padchest-gr/
Unsupervised Training of Neural Cellular Automata on Edge Devices
Jul 25, 2024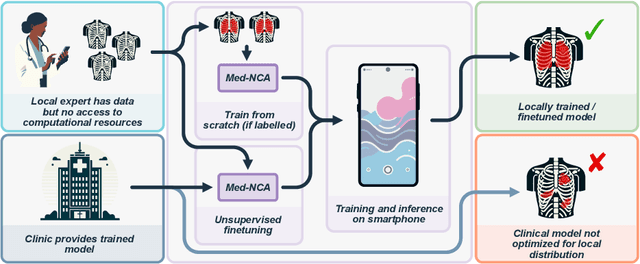
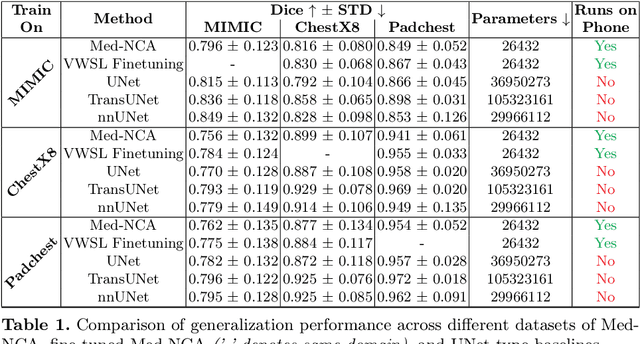
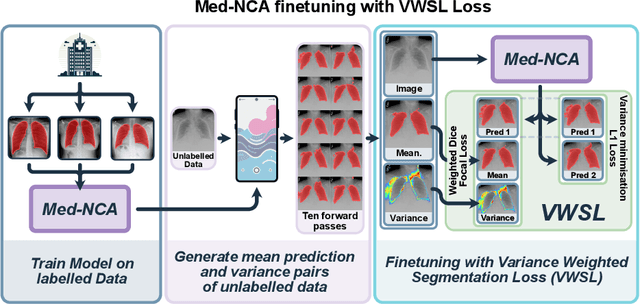
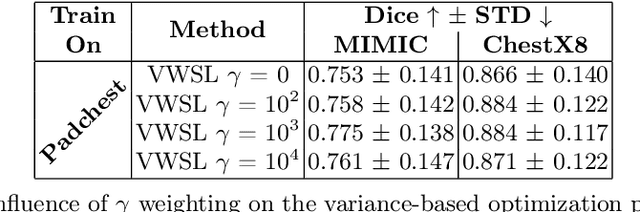
The disparity in access to machine learning tools for medical imaging across different regions significantly limits the potential for universal healthcare innovation, particularly in remote areas. Our research addresses this issue by implementing Neural Cellular Automata (NCA) training directly on smartphones for accessible X-ray lung segmentation. We confirm the practicality and feasibility of deploying and training these advanced models on five Android devices, improving medical diagnostics accessibility and bridging the tech divide to extend machine learning benefits in medical imaging to low- and middle-income countries (LMICs). We further enhance this approach with an unsupervised adaptation method using the novel Variance-Weighted Segmentation Loss (VWSL), which efficiently learns from unlabeled data by minimizing the variance from multiple NCA predictions. This strategy notably improves model adaptability and performance across diverse medical imaging contexts without the need for extensive computational resources or labeled datasets, effectively lowering the participation threshold. Our methodology, tested on three multisite X-ray datasets -- Padchest, ChestX-ray8, and MIMIC-III -- demonstrates improvements in segmentation Dice accuracy by 0.7 to 2.8%, compared to the classic Med-NCA. Additionally, in extreme cases where no digital copy is available and images must be captured by a phone from an X-ray lightbox or monitor, VWSL enhances Dice accuracy by 5-20%, demonstrating the method's robustness even with suboptimal image sources.
Augmenting Chest X-ray Datasets with Non-Expert Annotations
Sep 05, 2023The advancement of machine learning algorithms in medical image analysis requires the expansion of training datasets. A popular and cost-effective approach is automated annotation extraction from free-text medical reports, primarily due to the high costs associated with expert clinicians annotating chest X-ray images. However, it has been shown that the resulting datasets are susceptible to biases and shortcuts. Another strategy to increase the size of a dataset is crowdsourcing, a widely adopted practice in general computer vision with some success in medical image analysis. In a similar vein to crowdsourcing, we enhance two publicly available chest X-ray datasets by incorporating non-expert annotations. However, instead of using diagnostic labels, we annotate shortcuts in the form of tubes. We collect 3.5k chest drain annotations for CXR14, and 1k annotations for 4 different tube types in PadChest. We train a chest drain detector with the non-expert annotations that generalizes well to expert labels. Moreover, we compare our annotations to those provided by experts and show "moderate" to "almost perfect" agreement. Finally, we present a pathology agreement study to raise awareness about ground truth annotations. We make our annotations and code available.
CheXmask: a large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images
Jul 06, 2023The development of successful artificial intelligence models for chest X-ray analysis relies on large, diverse datasets with high-quality annotations. While several databases of chest X-ray images have been released, most include disease diagnosis labels but lack detailed pixel-level anatomical segmentation labels. To address this gap, we introduce an extensive chest X-ray multi-center segmentation dataset with uniform and fine-grain anatomical annotations for images coming from six well-known publicly available databases: CANDID-PTX, ChestX-ray8, Chexpert, MIMIC-CXR-JPG, Padchest, and VinDr-CXR, resulting in 676,803 segmentation masks. Our methodology utilizes the HybridGNet model to ensure consistent and high-quality segmentations across all datasets. Rigorous validation, including expert physician evaluation and automatic quality control, was conducted to validate the resulting masks. Additionally, we provide individualized quality indices per mask and an overall quality estimation per dataset. This dataset serves as a valuable resource for the broader scientific community, streamlining the development and assessment of innovative methodologies in chest X-ray analysis. The CheXmask dataset is publicly available at: \url{https://physionet.org/content/chexmask-cxr-segmentation-data/}.
Explainable Knowledge Distillation for On-device Chest X-Ray Classification
May 10, 2023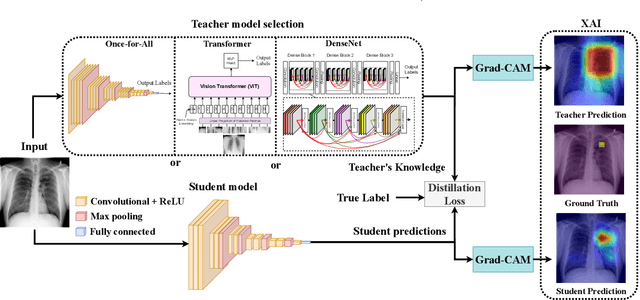
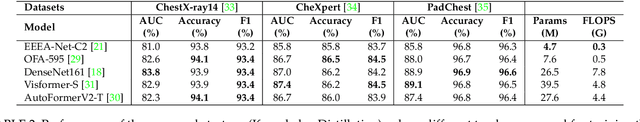
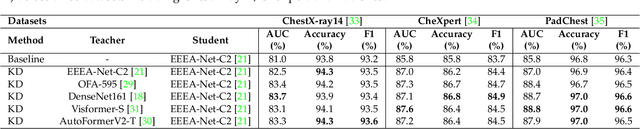
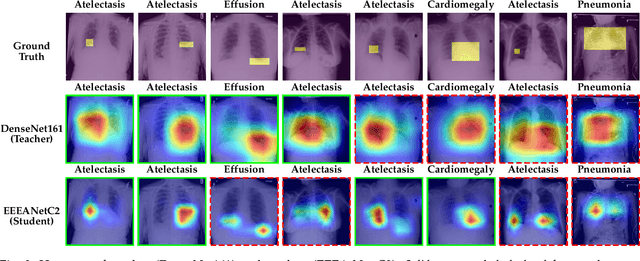
Automated multi-label chest X-rays (CXR) image classification has achieved substantial progress in clinical diagnosis via utilizing sophisticated deep learning approaches. However, most deep models have high computational demands, which makes them less feasible for compact devices with low computational requirements. To overcome this problem, we propose a knowledge distillation (KD) strategy to create the compact deep learning model for the real-time multi-label CXR image classification. We study different alternatives of CNNs and Transforms as the teacher to distill the knowledge to a smaller student. Then, we employed explainable artificial intelligence (XAI) to provide the visual explanation for the model decision improved by the KD. Our results on three benchmark CXR datasets show that our KD strategy provides the improved performance on the compact student model, thus being the feasible choice for many limited hardware platforms. For instance, when using DenseNet161 as the teacher network, EEEA-Net-C2 achieved an AUC of 83.7%, 87.1%, and 88.7% on the ChestX-ray14, CheXpert, and PadChest datasets, respectively, with fewer parameters of 4.7 million and computational cost of 0.3 billion FLOPS.
Deep learning for understanding multilabel imbalanced Chest X-ray datasets
Jul 28, 2022



Over the last few years, convolutional neural networks (CNNs) have dominated the field of computer vision thanks to their ability to extract features and their outstanding performance in classification problems, for example in the automatic analysis of X-rays. Unfortunately, these neural networks are considered black-box algorithms, i.e. it is impossible to understand how the algorithm has achieved the final result. To apply these algorithms in different fields and test how the methodology works, we need to use eXplainable AI techniques. Most of the work in the medical field focuses on binary or multiclass classification problems. However, in many real-life situations, such as chest X-rays, radiological signs of different diseases can appear at the same time. This gives rise to what is known as "multilabel classification problems". A disadvantage of these tasks is class imbalance, i.e. different labels do not have the same number of samples. The main contribution of this paper is a Deep Learning methodology for imbalanced, multilabel chest X-ray datasets. It establishes a baseline for the currently underutilised PadChest dataset and a new eXplainable AI technique based on heatmaps. This technique also includes probabilities and inter-model matching. The results of our system are promising, especially considering the number of labels used. Furthermore, the heatmaps match the expected areas, i.e. they mark the areas that an expert would use to make the decision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge