Yih Chung Tham
Generalist versus Specialist Vision Foundation Models for Ocular Disease and Oculomics
Sep 03, 2025Abstract:Medical foundation models, pre-trained with large-scale clinical data, demonstrate strong performance in diverse clinically relevant applications. RETFound, trained on nearly one million retinal images, exemplifies this approach in applications with retinal images. However, the emergence of increasingly powerful and multifold larger generalist foundation models such as DINOv2 and DINOv3 raises the question of whether domain-specific pre-training remains essential, and if so, what gap persists. To investigate this, we systematically evaluated the adaptability of DINOv2 and DINOv3 in retinal image applications, compared to two specialist RETFound models, RETFound-MAE and RETFound-DINOv2. We assessed performance on ocular disease detection and systemic disease prediction using two adaptation strategies: fine-tuning and linear probing. Data efficiency and adaptation efficiency were further analysed to characterise trade-offs between predictive performance and computational cost. Our results show that although scaling generalist models yields strong adaptability across diverse tasks, RETFound-DINOv2 consistently outperforms these generalist foundation models in ocular-disease detection and oculomics tasks, demonstrating stronger generalisability and data efficiency. These findings suggest that specialist retinal foundation models remain the most effective choice for clinical applications, while the narrowing gap with generalist foundation models suggests that continued data and model scaling can deliver domain-relevant gains and position them as strong foundations for future medical foundation models.
Performance of GPT-5 Frontier Models in Ophthalmology Question Answering
Aug 14, 2025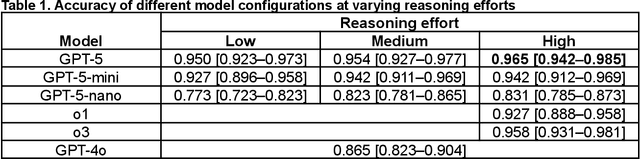
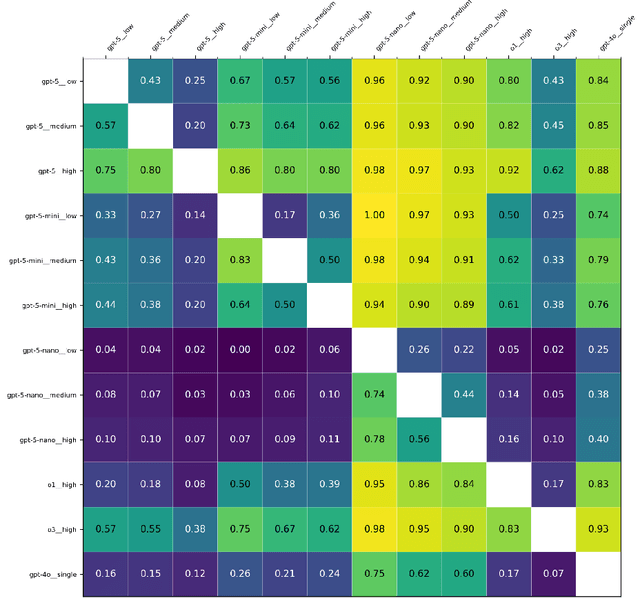
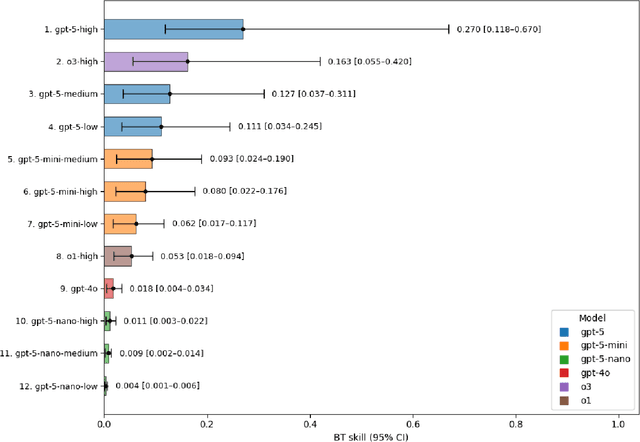
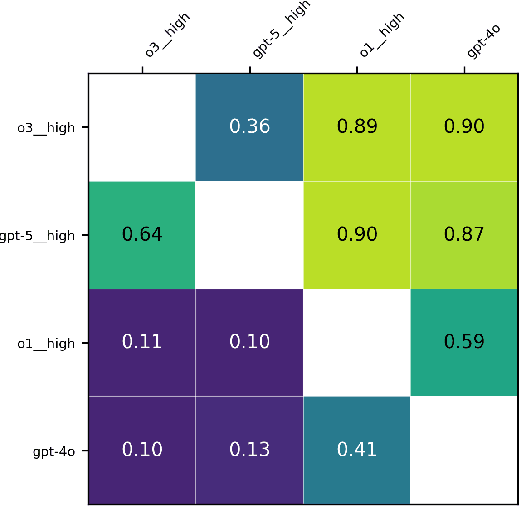
Abstract:Large language models (LLMs) such as GPT-5 integrate advanced reasoning capabilities that may improve performance on complex medical question-answering tasks. For this latest generation of reasoning models, the configurations that maximize both accuracy and cost-efficiency have yet to be established. We evaluated 12 configurations of OpenAI's GPT-5 series (three model tiers across four reasoning effort settings) alongside o1-high, o3-high, and GPT-4o, using 260 closed-access multiple-choice questions from the American Academy of Ophthalmology Basic Clinical Science Course (BCSC) dataset. The primary outcome was multiple-choice accuracy; secondary outcomes included head-to-head ranking via a Bradley-Terry model, rationale quality assessment using a reference-anchored, pairwise LLM-as-a-judge framework, and analysis of accuracy-cost trade-offs using token-based cost estimates. GPT-5-high achieved the highest accuracy (0.965; 95% CI, 0.942-0.985), outperforming all GPT-5-nano variants (P < .001), o1-high (P = .04), and GPT-4o (P < .001), but not o3-high (0.958; 95% CI, 0.931-0.981). GPT-5-high ranked first in both accuracy (1.66x stronger than o3-high) and rationale quality (1.11x stronger than o3-high). Cost-accuracy analysis identified several GPT-5 configurations on the Pareto frontier, with GPT-5-mini-low offering the most favorable low-cost, high-performance balance. These results benchmark GPT-5 on a high-quality ophthalmology dataset, demonstrate the influence of reasoning effort on accuracy, and introduce an autograder framework for scalable evaluation of LLM-generated answers against reference standards in ophthalmology.
An integrated language-vision foundation model for conversational diagnostics and triaging in primary eye care
May 13, 2025Abstract:Current deep learning models are mostly task specific and lack a user-friendly interface to operate. We present Meta-EyeFM, a multi-function foundation model that integrates a large language model (LLM) with vision foundation models (VFMs) for ocular disease assessment. Meta-EyeFM leverages a routing mechanism to enable accurate task-specific analysis based on text queries. Using Low Rank Adaptation, we fine-tuned our VFMs to detect ocular and systemic diseases, differentiate ocular disease severity, and identify common ocular signs. The model achieved 100% accuracy in routing fundus images to appropriate VFMs, which achieved $\ge$ 82.2% accuracy in disease detection, $\ge$ 89% in severity differentiation, $\ge$ 76% in sign identification. Meta-EyeFM was 11% to 43% more accurate than Gemini-1.5-flash and ChatGPT-4o LMMs in detecting various eye diseases and comparable to an ophthalmologist. This system offers enhanced usability and diagnostic performance, making it a valuable decision support tool for primary eye care or an online LLM for fundus evaluation.
A Clinician-Friendly Platform for Ophthalmic Image Analysis Without Technical Barriers
Apr 22, 2025Abstract:Artificial intelligence (AI) shows remarkable potential in medical imaging diagnostics, but current models typically require retraining when deployed across different clinical centers, limiting their widespread adoption. We introduce GlobeReady, a clinician-friendly AI platform that enables ocular disease diagnosis without retraining/fine-tuning or technical expertise. GlobeReady achieves high accuracy across imaging modalities: 93.9-98.5% for an 11-category fundus photo dataset and 87.2-92.7% for a 15-category OCT dataset. Through training-free local feature augmentation, it addresses domain shifts across centers and populations, reaching an average accuracy of 88.9% across five centers in China, 86.3% in Vietnam, and 90.2% in the UK. The built-in confidence-quantifiable diagnostic approach further boosted accuracy to 94.9-99.4% (fundus) and 88.2-96.2% (OCT), while identifying out-of-distribution cases at 86.3% (49 CFP categories) and 90.6% (13 OCT categories). Clinicians from multiple countries rated GlobeReady highly (average 4.6 out of 5) for its usability and clinical relevance. These results demonstrate GlobeReady's robust, scalable diagnostic capability and potential to support ophthalmic care without technical barriers.
Benchmarking Next-Generation Reasoning-Focused Large Language Models in Ophthalmology: A Head-to-Head Evaluation on 5,888 Items
Apr 15, 2025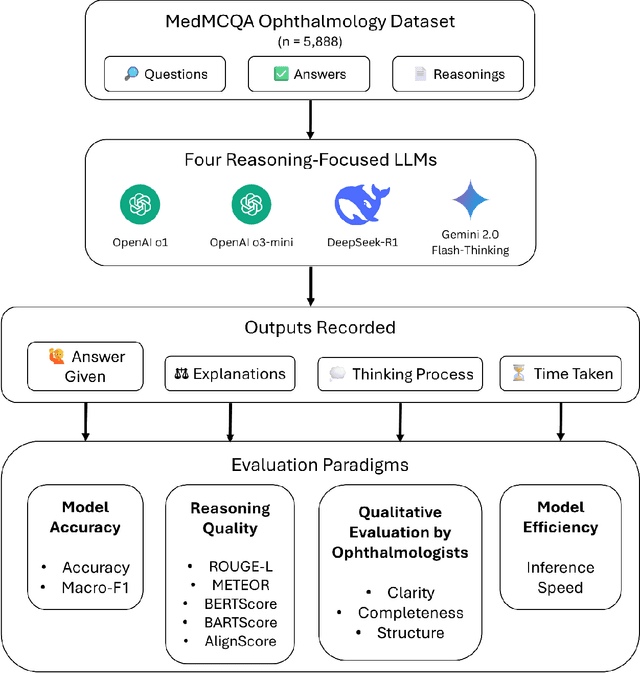
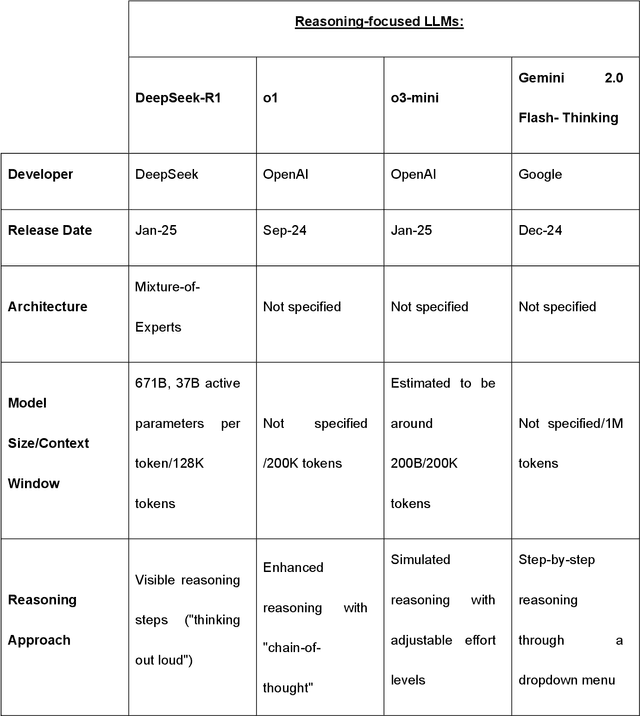
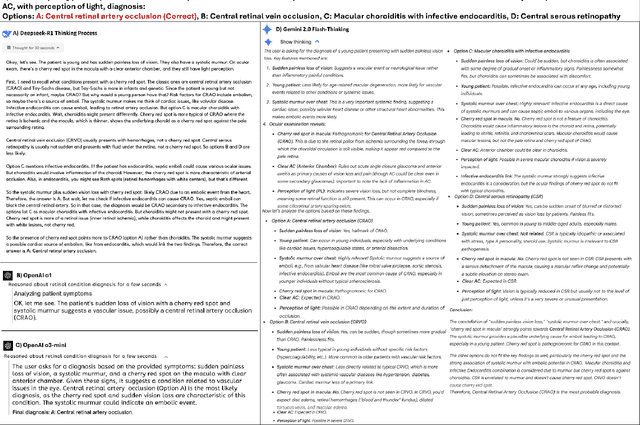
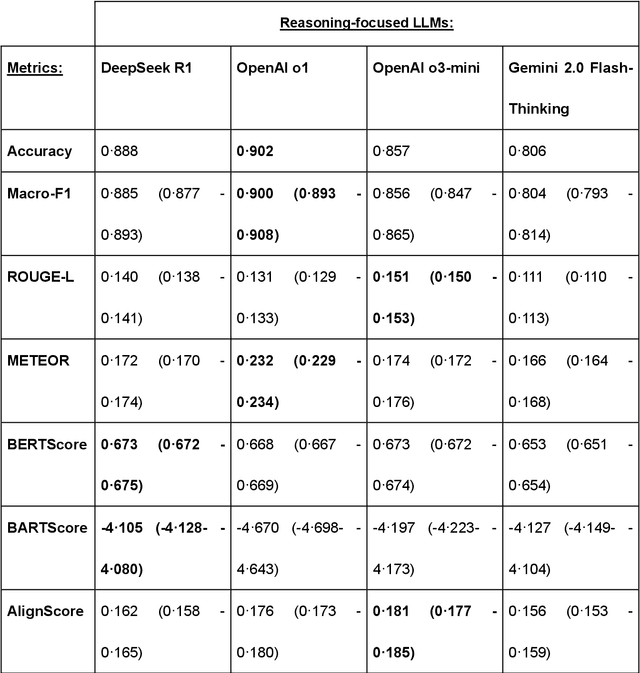
Abstract:Recent advances in reasoning-focused large language models (LLMs) mark a shift from general LLMs toward models designed for complex decision-making, a crucial aspect in medicine. However, their performance in specialized domains like ophthalmology remains underexplored. This study comprehensively evaluated and compared the accuracy and reasoning capabilities of four newly developed reasoning-focused LLMs, namely DeepSeek-R1, OpenAI o1, o3-mini, and Gemini 2.0 Flash-Thinking. Each model was assessed using 5,888 multiple-choice ophthalmology exam questions from the MedMCQA dataset in zero-shot setting. Quantitative evaluation included accuracy, Macro-F1, and five text-generation metrics (ROUGE-L, METEOR, BERTScore, BARTScore, and AlignScore), computed against ground-truth reasonings. Average inference time was recorded for a subset of 100 randomly selected questions. Additionally, two board-certified ophthalmologists qualitatively assessed clarity, completeness, and reasoning structure of responses to differential diagnosis questions.O1 (0.902) and DeepSeek-R1 (0.888) achieved the highest accuracy, with o1 also leading in Macro-F1 (0.900). The performance of models across the text-generation metrics varied: O3-mini excelled in ROUGE-L (0.151), o1 in METEOR (0.232), DeepSeek-R1 and o3-mini tied for BERTScore (0.673), DeepSeek-R1 (-4.105) and Gemini 2.0 Flash-Thinking (-4.127) performed best in BARTScore, while o3-mini (0.181) and o1 (0.176) led AlignScore. Inference time across the models varied, with DeepSeek-R1 being slowest (40.4 seconds) and Gemini 2.0 Flash-Thinking fastest (6.7 seconds). Qualitative evaluation revealed that DeepSeek-R1 and Gemini 2.0 Flash-Thinking tended to provide detailed and comprehensive intermediate reasoning, whereas o1 and o3-mini displayed concise and summarized justifications.
Is an Ultra Large Natural Image-Based Foundation Model Superior to a Retina-Specific Model for Detecting Ocular and Systemic Diseases?
Feb 10, 2025Abstract:The advent of foundation models (FMs) is transforming medical domain. In ophthalmology, RETFound, a retina-specific FM pre-trained sequentially on 1.4 million natural images and 1.6 million retinal images, has demonstrated high adaptability across clinical applications. Conversely, DINOv2, a general-purpose vision FM pre-trained on 142 million natural images, has shown promise in non-medical domains. However, its applicability to clinical tasks remains underexplored. To address this, we conducted head-to-head evaluations by fine-tuning RETFound and three DINOv2 models (large, base, small) for ocular disease detection and systemic disease prediction tasks, across eight standardized open-source ocular datasets, as well as the Moorfields AlzEye and the UK Biobank datasets. DINOv2-large model outperformed RETFound in detecting diabetic retinopathy (AUROC=0.850-0.952 vs 0.823-0.944, across three datasets, all P<=0.007) and multi-class eye diseases (AUROC=0.892 vs. 0.846, P<0.001). In glaucoma, DINOv2-base model outperformed RETFound (AUROC=0.958 vs 0.940, P<0.001). Conversely, RETFound achieved superior performance over all DINOv2 models in predicting heart failure, myocardial infarction, and ischaemic stroke (AUROC=0.732-0.796 vs 0.663-0.771, all P<0.001). These trends persisted even with 10% of the fine-tuning data. These findings showcase the distinct scenarios where general-purpose and domain-specific FMs excel, highlighting the importance of aligning FM selection with task-specific requirements to optimise clinical performance.
Block Expanded DINORET: Adapting Natural Domain Foundation Models for Retinal Imaging Without Catastrophic Forgetting
Sep 25, 2024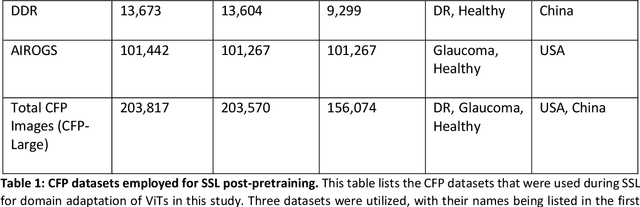
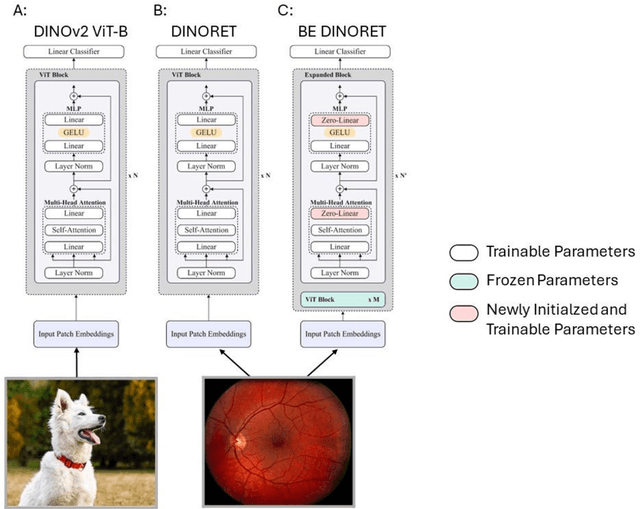
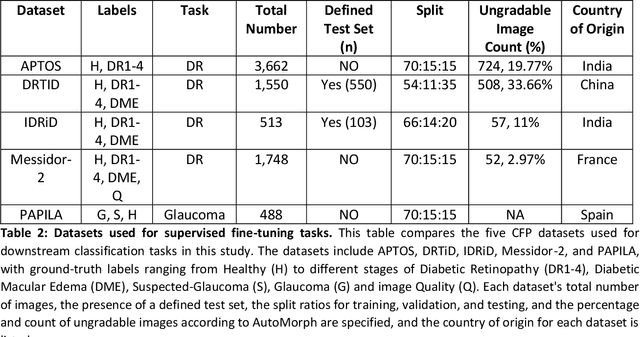
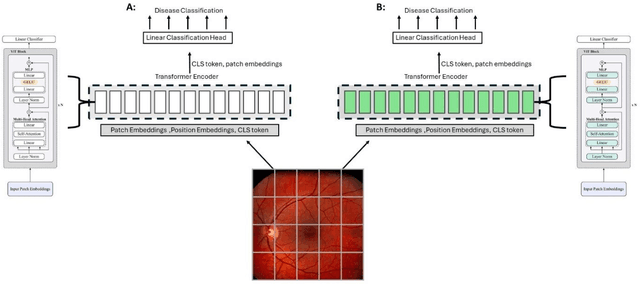
Abstract:Integrating deep learning into medical imaging is poised to greatly advance diagnostic methods but it faces challenges with generalizability. Foundation models, based on self-supervised learning, address these issues and improve data efficiency. Natural domain foundation models show promise for medical imaging, but systematic research evaluating domain adaptation, especially using self-supervised learning and parameter-efficient fine-tuning, remains underexplored. Additionally, little research addresses the issue of catastrophic forgetting during fine-tuning of foundation models. We adapted the DINOv2 vision transformer for retinal imaging classification tasks using self-supervised learning and generated two novel foundation models termed DINORET and BE DINORET. Publicly available color fundus photographs were employed for model development and subsequent fine-tuning for diabetic retinopathy staging and glaucoma detection. We introduced block expansion as a novel domain adaptation strategy and assessed the models for catastrophic forgetting. Models were benchmarked to RETFound, a state-of-the-art foundation model in ophthalmology. DINORET and BE DINORET demonstrated competitive performance on retinal imaging tasks, with the block expanded model achieving the highest scores on most datasets. Block expansion successfully mitigated catastrophic forgetting. Our few-shot learning studies indicated that DINORET and BE DINORET outperform RETFound in terms of data-efficiency. This study highlights the potential of adapting natural domain vision models to retinal imaging using self-supervised learning and block expansion. BE DINORET offers robust performance without sacrificing previously acquired capabilities. Our findings suggest that these methods could enable healthcare institutions to develop tailored vision models for their patient populations, enhancing global healthcare inclusivity.
Common and Rare Fundus Diseases Identification Using Vision-Language Foundation Model with Knowledge of Over 400 Diseases
Jun 13, 2024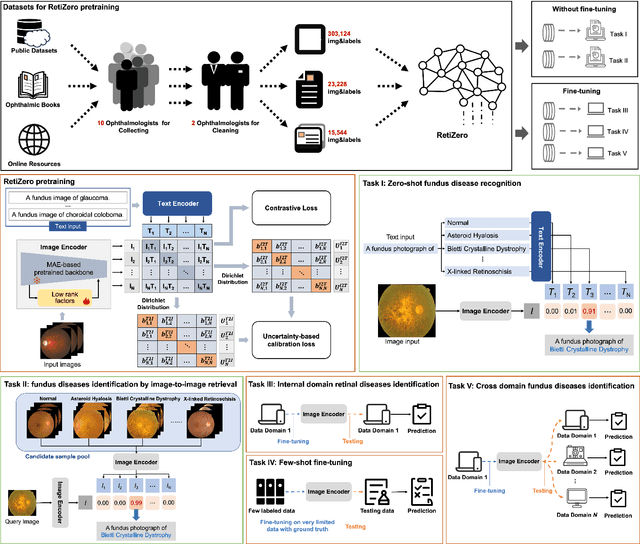
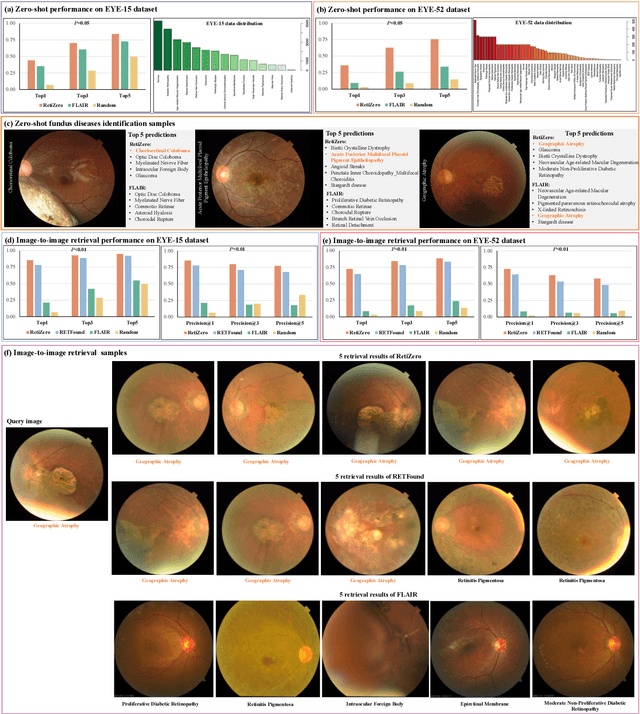
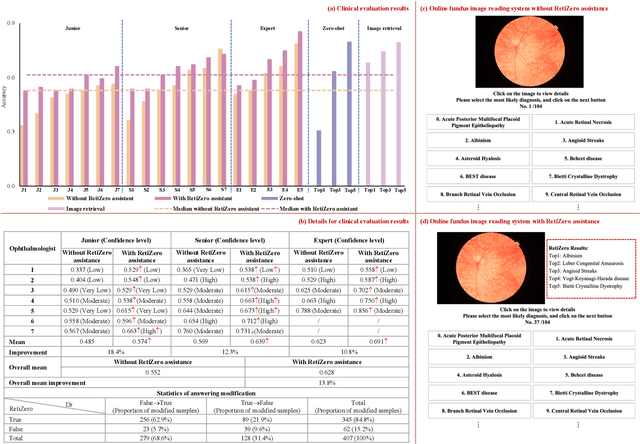
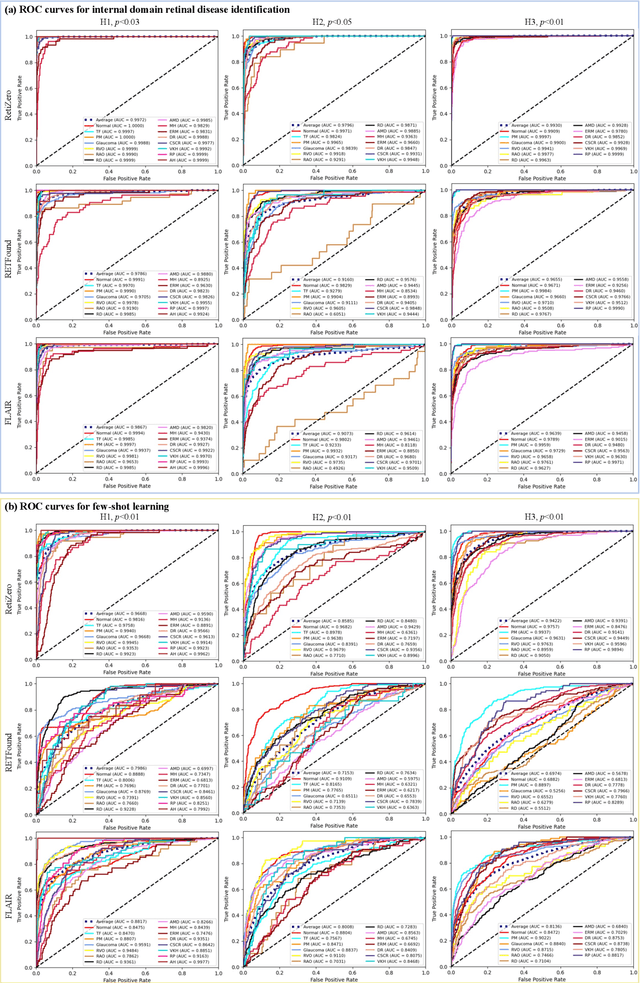
Abstract:The current retinal artificial intelligence models were trained using data with a limited category of diseases and limited knowledge. In this paper, we present a retinal vision-language foundation model (RetiZero) with knowledge of over 400 fundus diseases. Specifically, we collected 341,896 fundus images paired with text descriptions from 29 publicly available datasets, 180 ophthalmic books, and online resources, encompassing over 400 fundus diseases across multiple countries and ethnicities. RetiZero achieved outstanding performance across various downstream tasks, including zero-shot retinal disease recognition, image-to-image retrieval, internal domain and cross-domain retinal disease classification, and few-shot fine-tuning. Specially, in the zero-shot scenario, RetiZero achieved a Top5 score of 0.8430 and 0.7561 on 15 and 52 fundus diseases respectively. In the image-retrieval task, RetiZero achieved a Top5 score of 0.9500 and 0.8860 on 15 and 52 retinal diseases respectively. Furthermore, clinical evaluations by ophthalmology experts from different countries demonstrate that RetiZero can achieve performance comparable to experienced ophthalmologists using zero-shot and image retrieval methods without requiring model retraining. These capabilities of retinal disease identification strengthen our RetiZero foundation model in clinical implementation.
EyeFound: A Multimodal Generalist Foundation Model for Ophthalmic Imaging
May 22, 2024Abstract:Artificial intelligence (AI) is vital in ophthalmology, tackling tasks like diagnosis, classification, and visual question answering (VQA). However, existing AI models in this domain often require extensive annotation and are task-specific, limiting their clinical utility. While recent developments have brought about foundation models for ophthalmology, they are limited by the need to train separate weights for each imaging modality, preventing a comprehensive representation of multi-modal features. This highlights the need for versatile foundation models capable of handling various tasks and modalities in ophthalmology. To address this gap, we present EyeFound, a multimodal foundation model for ophthalmic images. Unlike existing models, EyeFound learns generalizable representations from unlabeled multimodal retinal images, enabling efficient model adaptation across multiple applications. Trained on 2.78 million images from 227 hospitals across 11 ophthalmic modalities, EyeFound facilitates generalist representations and diverse multimodal downstream tasks, even for detecting challenging rare diseases. It outperforms previous work RETFound in diagnosing eye diseases, predicting systemic disease incidents, and zero-shot multimodal VQA. EyeFound provides a generalizable solution to improve model performance and lessen the annotation burden on experts, facilitating widespread clinical AI applications for retinal imaging.
A Clinical-oriented Multi-level Contrastive Learning Method for Disease Diagnosis in Low-quality Medical Images
Apr 07, 2024Abstract:Representation learning offers a conduit to elucidate distinctive features within the latent space and interpret the deep models. However, the randomness of lesion distribution and the complexity of low-quality factors in medical images pose great challenges for models to extract key lesion features. Disease diagnosis methods guided by contrastive learning (CL) have shown significant advantages in lesion feature representation. Nevertheless, the effectiveness of CL is highly dependent on the quality of the positive and negative sample pairs. In this work, we propose a clinical-oriented multi-level CL framework that aims to enhance the model's capacity to extract lesion features and discriminate between lesion and low-quality factors, thereby enabling more accurate disease diagnosis from low-quality medical images. Specifically, we first construct multi-level positive and negative pairs to enhance the model's comprehensive recognition capability of lesion features by integrating information from different levels and qualities of medical images. Moreover, to improve the quality of the learned lesion embeddings, we introduce a dynamic hard sample mining method based on self-paced learning. The proposed CL framework is validated on two public medical image datasets, EyeQ and Chest X-ray, demonstrating superior performance compared to other state-of-the-art disease diagnostic methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge