Weiguo Lu
Exploiting DINOv3-Based Self-Supervised Features for Robust Few-Shot Medical Image Segmentation
Jan 12, 2026Abstract:Deep learning-based automatic medical image segmentation plays a critical role in clinical diagnosis and treatment planning but remains challenging in few-shot scenarios due to the scarcity of annotated training data. Recently, self-supervised foundation models such as DINOv3, which were trained on large natural image datasets, have shown strong potential for dense feature extraction that can help with the few-shot learning challenge. Yet, their direct application to medical images is hindered by domain differences. In this work, we propose DINO-AugSeg, a novel framework that leverages DINOv3 features to address the few-shot medical image segmentation challenge. Specifically, we introduce WT-Aug, a wavelet-based feature-level augmentation module that enriches the diversity of DINOv3-extracted features by perturbing frequency components, and CG-Fuse, a contextual information-guided fusion module that exploits cross-attention to integrate semantic-rich low-resolution features with spatially detailed high-resolution features. Extensive experiments on six public benchmarks spanning five imaging modalities, including MRI, CT, ultrasound, endoscopy, and dermoscopy, demonstrate that DINO-AugSeg consistently outperforms existing methods under limited-sample conditions. The results highlight the effectiveness of incorporating wavelet-domain augmentation and contextual fusion for robust feature representation, suggesting DINO-AugSeg as a promising direction for advancing few-shot medical image segmentation. Code and data will be made available on https://github.com/apple1986/DINO-AugSeg.
A SAM-guided and Match-based Semi-Supervised Segmentation Framework for Medical Imaging
Nov 25, 2024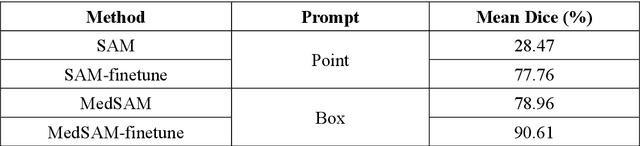
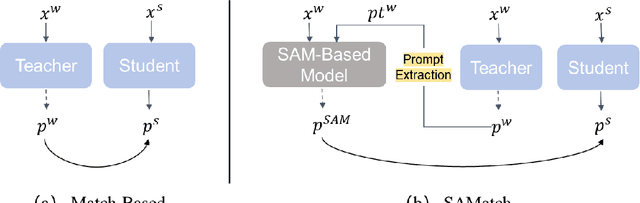
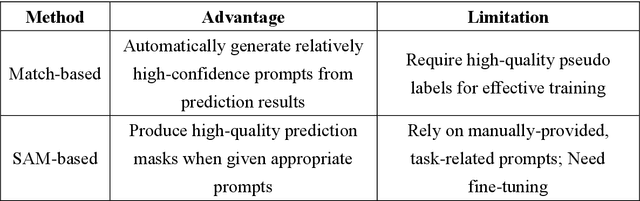
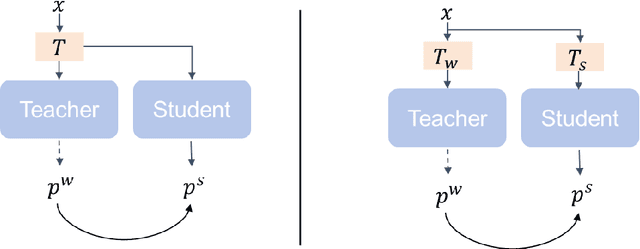
Abstract:This study introduces SAMatch, a SAM-guided Match-based framework for semi-supervised medical image segmentation, aimed at improving pseudo label quality in data-scarce scenarios. While Match-based frameworks are effective, they struggle with low-quality pseudo labels due to the absence of ground truth. SAM, pre-trained on a large dataset, generalizes well across diverse tasks and assists in generating high-confidence prompts, which are then used to refine pseudo labels via fine-tuned SAM. SAMatch is trained end-to-end, allowing for dynamic interaction between the models. Experiments on the ACDC cardiac MRI, BUSI breast ultrasound, and MRLiver datasets show SAMatch achieving state-of-the-art results, with Dice scores of 89.36%, 77.76%, and 80.04%, respectively, using minimal labeled data. SAMatch effectively addresses challenges in semi-supervised segmentation, offering a powerful tool for segmentation in data-limited environments. Code and data are available at https://github.com/apple1986/SAMatch.
Diffusion Model Conditioning on Gaussian Mixture Model and Negative Gaussian Mixture Gradient
Feb 01, 2024



Abstract:Diffusion models (DMs) are a type of generative model that has a huge impact on image synthesis and beyond. They achieve state-of-the-art generation results in various generative tasks. A great diversity of conditioning inputs, such as text or bounding boxes, are accessible to control the generation. In this work, we propose a conditioning mechanism utilizing Gaussian mixture models (GMMs) as feature conditioning to guide the denoising process. Based on set theory, we provide a comprehensive theoretical analysis that shows that conditional latent distribution based on features and classes is significantly different, so that conditional latent distribution on features produces fewer defect generations than conditioning on classes. Two diffusion models conditioned on the Gaussian mixture model are trained separately for comparison. Experiments support our findings. A novel gradient function called the negative Gaussian mixture gradient (NGMG) is proposed and applied in diffusion model training with an additional classifier. Training stability has improved. We also theoretically prove that NGMG shares the same benefit as the Earth Mover distance (Wasserstein) as a more sensible cost function when learning distributions supported by low-dimensional manifolds.
An Efficient 1 Iteration Learning Algorithm for Gaussian Mixture Model And Gaussian Mixture Embedding For Neural Network
Sep 06, 2023



Abstract:We propose an Gaussian Mixture Model (GMM) learning algorithm, based on our previous work of GMM expansion idea. The new algorithm brings more robustness and simplicity than classic Expectation Maximization (EM) algorithm. It also improves the accuracy and only take 1 iteration for learning. We theoretically proof that this new algorithm is guarantee to converge regardless the parameters initialisation. We compare our GMM expansion method with classic probability layers in neural network leads to demonstrably better capability to overcome data uncertainty and inverse problem. Finally, we test GMM based generator which shows a potential to build further application that able to utilized distribution random sampling for stochastic variation as well as variation control.
Recurrence-free Survival Prediction under the Guidance of Automatic Gross Tumor Volume Segmentation for Head and Neck Cancers
Sep 22, 2022



Abstract:For Head and Neck Cancers (HNC) patient management, automatic gross tumor volume (GTV) segmentation and accurate pre-treatment cancer recurrence prediction are of great importance to assist physicians in designing personalized management plans, which have the potential to improve the treatment outcome and quality of life for HNC patients. In this paper, we developed an automated primary tumor (GTVp) and lymph nodes (GTVn) segmentation method based on combined pre-treatment positron emission tomography/computed tomography (PET/CT) scans of HNC patients. We extracted radiomics features from the segmented tumor volume and constructed a multi-modality tumor recurrence-free survival (RFS) prediction model, which fused the prediction results from separate CT radiomics, PET radiomics, and clinical models. We performed 5-fold cross-validation to train and evaluate our methods on the MICCAI 2022 HEad and neCK TumOR segmentation and outcome prediction challenge (HECKTOR) dataset. The ensemble prediction results on the testing cohort achieved Dice scores of 0.77 and 0.73 for GTVp and GTVn segmentation, respectively, and a C-index value of 0.67 for RFS prediction. The code is publicly available (https://github.com/wangkaiwan/HECKTOR-2022-AIRT). Our team's name is AIRT.
Leveraging Global Binary Masks for Structure Segmentation in Medical Images
May 13, 2022



Abstract:Deep learning (DL) models for medical image segmentation are highly influenced by intensity variations of input images and lack generalization due to primarily utilizing pixels' intensity information for inference. Acquiring sufficient training data is another challenge limiting models' applications. We proposed to leverage the consistency of organs' anatomical shape and position information in medical images. We introduced a framework leveraging recurring anatomical patterns through global binary masks for organ segmentation. Two scenarios were studied.1) Global binary masks were the only model's (i.e. U-Net) input, forcing exclusively encoding organs' position and shape information for segmentation/localization.2) Global binary masks were incorporated as an additional channel functioning as position/shape clues to mitigate training data scarcity. Two datasets of the brain and heart CT images with their ground-truth were split into (26:10:10) and (12:3:5) for training, validation, and test respectively. Training exclusively on global binary masks led to Dice scores of 0.77(0.06) and 0.85(0.04), with the average Euclidian distance of 3.12(1.43)mm and 2.5(0.93)mm relative to the center of mass of the ground truth for the brain and heart structures respectively. The outcomes indicate that a surprising degree of position and shape information is encoded through global binary masks. Incorporating global binary masks led to significantly higher accuracy relative to the model trained on only CT images in small subsets of training data; the performance improved by 4.3-125.3% and 1.3-48.1% for 1-8 training cases of the brain and heart datasets respectively. The findings imply the advantages of utilizing global binary masks for building generalizable models and to compensate for training data scarcity.
Registration-Guided Deep Learning Image Segmentation for Cone Beam CT-based Online Adaptive Radiotherapy
Aug 19, 2021



Abstract:Adaptive radiotherapy (ART), especially online ART, effectively accounts for positioning errors and anatomical changes. One key component of online ART is accurately and efficiently delineating organs at risk (OARs) and targets on online images, such as CBCT, to meet the online demands of plan evaluation and adaptation. Deep learning (DL)-based automatic segmentation has gained great success in segmenting planning CT, but its applications to CBCT yielded inferior results due to the low image quality and limited available contour labels for training. To overcome these obstacles to online CBCT segmentation, we propose a registration-guided DL (RgDL) segmentation framework that integrates image registration algorithms and DL segmentation models. The registration algorithm generates initial contours, which were used as guidance by DL model to obtain accurate final segmentations. We had two implementations the proposed framework--Rig-RgDL (Rig for rigid body) and Def-RgDL (Def for deformable)--with rigid body (RB) registration or deformable image registration (DIR) as the registration algorithm respectively and U-Net as DL model architecture. The two implementations of RgDL framework were trained and evaluated on seven OARs in an institutional clinical Head and Neck (HN) dataset. Compared to the baseline approaches using the registration or the DL alone, RgDL achieved more accurate segmentation, as measured by higher mean Dice similarity coefficients (DSC) and other distance-based metrics. Rig-RgDL achieved a DSC of 84.5% on seven OARs on average, higher than RB or DL alone by 4.5% and 4.7%. The DSC of Def-RgDL is 86.5%, higher than DIR or DL alone by 2.4% and 6.7%. The inference time took by the DL model to generate final segmentations of seven OARs is less than one second in RgDL. The resulting segmentation accuracy and efficiency show the promise of applying RgDL framework for online ART.
Saliency-Guided Deep Learning Network for Automatic Tumor Bed Volume Delineation in Post-operative Breast Irradiation
May 06, 2021



Abstract:Efficient, reliable and reproducible target volume delineation is a key step in the effective planning of breast radiotherapy. However, post-operative breast target delineation is challenging as the contrast between the tumor bed volume (TBV) and normal breast tissue is relatively low in CT images. In this study, we propose to mimic the marker-guidance procedure in manual target delineation. We developed a saliency-based deep learning segmentation (SDL-Seg) algorithm for accurate TBV segmentation in post-operative breast irradiation. The SDL-Seg algorithm incorporates saliency information in the form of markers' location cues into a U-Net model. The design forces the model to encode the location-related features, which underscores regions with high saliency levels and suppresses low saliency regions. The saliency maps were generated by identifying markers on CT images. Markers' locations were then converted to probability maps using a distance-transformation coupled with a Gaussian filter. Subsequently, the CT images and the corresponding saliency maps formed a multi-channel input for the SDL-Seg network. Our in-house dataset was comprised of 145 prone CT images from 29 post-operative breast cancer patients, who received 5-fraction partial breast irradiation (PBI) regimen on GammaPod. The performance of the proposed method was compared against basic U-Net. Our model achieved mean (standard deviation) of 76.4 %, 6.76 mm, and 1.9 mm for DSC, HD95, and ASD respectively on the test set with computation time of below 11 seconds per one CT volume. SDL-Seg showed superior performance relative to basic U-Net for all the evaluation metrics while preserving low computation cost. The findings demonstrate that SDL-Seg is a promising approach for improving the efficiency and accuracy of the on-line treatment planning procedure of PBI, such as GammaPod based PBI.
Breast Ultrasound Computer-Aided Diagnosis Using Structure-Aware Triplet Path Networks
Aug 09, 2019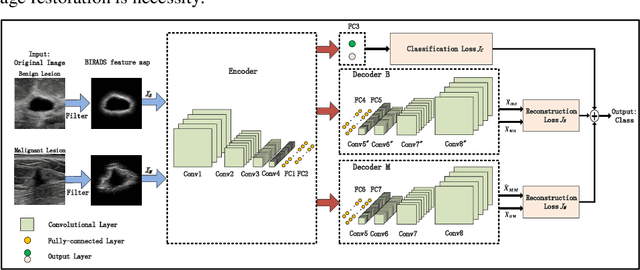
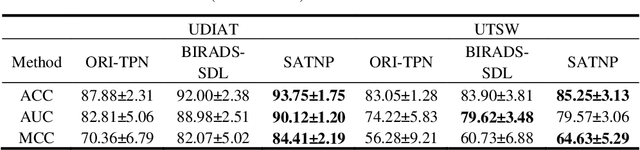
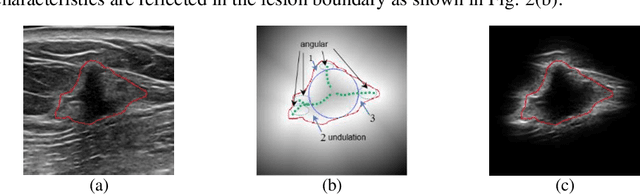
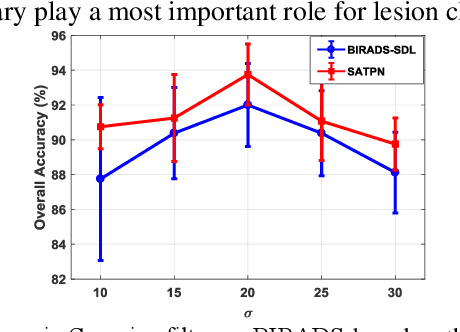
Abstract:Breast ultrasound (US) is an effective imaging modality for breast cancer detec-tion and diagnosis. The structural characteristics of breast lesion play an im-portant role in Computer-Aided Diagnosis (CAD). In this paper, a novel struc-ture-aware triplet path networks (SATPN) was designed to integrate classifica-tion and two image reconstruction tasks to achieve accurate diagnosis on US im-ages with small training dataset. Specifically, we enhance clinically-approved breast lesion structure characteristics though converting original breast US imag-es to BIRADS-oriented feature maps (BFMs) with a distance-transformation coupled Gaussian filter. Then, the converted BFMs were used as the inputs of SATPN, which performed lesion classification task and two unsupervised stacked convolutional Auto-Encoder (SCAE) networks for benign and malignant image reconstruction tasks, independently. We trained the SATPN with an alter-native learning strategy by balancing image reconstruction error and classification label prediction error. At the test stage, the lesion label was determined by the weighted voting with reconstruction error and label prediction error. We com-pared the performance of the SATPN with TPN using original image as input and our previous developed semi-supervised deep learning methods using BFMs as inputs. Experimental results on two breast US datasets showed that SATPN ranked the best among the three networks, with classification accuracy around 93.5%. These findings indicated that SATPN is promising for effective breast US lesion CAD using small datasets.
Three-Dimensional Dose Prediction for Lung IMRT Patients with Deep Neural Networks: Robust Learning from Heterogeneous Beam Configurations
Dec 17, 2018
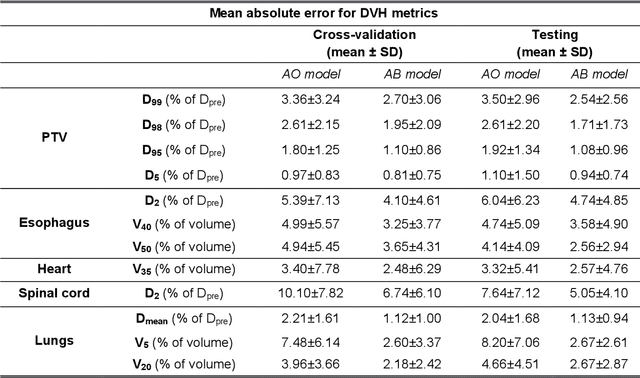
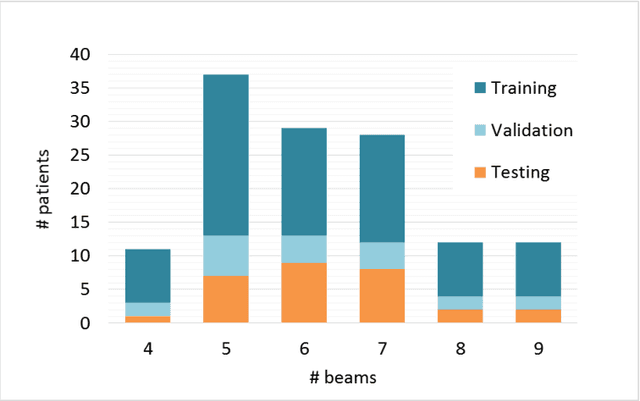
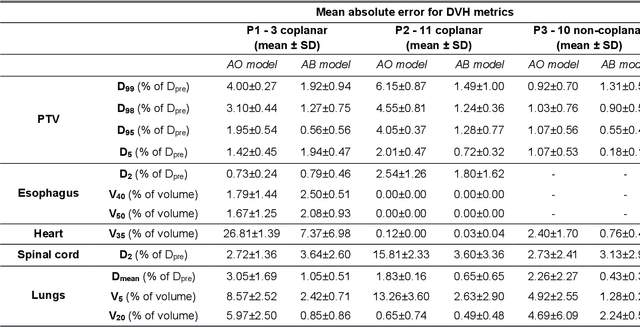
Abstract:The use of neural networks to directly predict three-dimensional dose distributions for automatic planning is becoming popular. However, the existing methods only use patient anatomy as input and assume consistent beam configuration for all patients in the training database. The purpose of this work is to develop a more general model that, in addition to patient anatomy, also considers variable beam configurations, to achieve a more comprehensive automatic planning with a potentially easier clinical implementation, without the need of training specific models for different beam settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge