Mingli Chen
Leveraging Global Binary Masks for Structure Segmentation in Medical Images
May 13, 2022



Abstract:Deep learning (DL) models for medical image segmentation are highly influenced by intensity variations of input images and lack generalization due to primarily utilizing pixels' intensity information for inference. Acquiring sufficient training data is another challenge limiting models' applications. We proposed to leverage the consistency of organs' anatomical shape and position information in medical images. We introduced a framework leveraging recurring anatomical patterns through global binary masks for organ segmentation. Two scenarios were studied.1) Global binary masks were the only model's (i.e. U-Net) input, forcing exclusively encoding organs' position and shape information for segmentation/localization.2) Global binary masks were incorporated as an additional channel functioning as position/shape clues to mitigate training data scarcity. Two datasets of the brain and heart CT images with their ground-truth were split into (26:10:10) and (12:3:5) for training, validation, and test respectively. Training exclusively on global binary masks led to Dice scores of 0.77(0.06) and 0.85(0.04), with the average Euclidian distance of 3.12(1.43)mm and 2.5(0.93)mm relative to the center of mass of the ground truth for the brain and heart structures respectively. The outcomes indicate that a surprising degree of position and shape information is encoded through global binary masks. Incorporating global binary masks led to significantly higher accuracy relative to the model trained on only CT images in small subsets of training data; the performance improved by 4.3-125.3% and 1.3-48.1% for 1-8 training cases of the brain and heart datasets respectively. The findings imply the advantages of utilizing global binary masks for building generalizable models and to compensate for training data scarcity.
Registration-Guided Deep Learning Image Segmentation for Cone Beam CT-based Online Adaptive Radiotherapy
Aug 19, 2021



Abstract:Adaptive radiotherapy (ART), especially online ART, effectively accounts for positioning errors and anatomical changes. One key component of online ART is accurately and efficiently delineating organs at risk (OARs) and targets on online images, such as CBCT, to meet the online demands of plan evaluation and adaptation. Deep learning (DL)-based automatic segmentation has gained great success in segmenting planning CT, but its applications to CBCT yielded inferior results due to the low image quality and limited available contour labels for training. To overcome these obstacles to online CBCT segmentation, we propose a registration-guided DL (RgDL) segmentation framework that integrates image registration algorithms and DL segmentation models. The registration algorithm generates initial contours, which were used as guidance by DL model to obtain accurate final segmentations. We had two implementations the proposed framework--Rig-RgDL (Rig for rigid body) and Def-RgDL (Def for deformable)--with rigid body (RB) registration or deformable image registration (DIR) as the registration algorithm respectively and U-Net as DL model architecture. The two implementations of RgDL framework were trained and evaluated on seven OARs in an institutional clinical Head and Neck (HN) dataset. Compared to the baseline approaches using the registration or the DL alone, RgDL achieved more accurate segmentation, as measured by higher mean Dice similarity coefficients (DSC) and other distance-based metrics. Rig-RgDL achieved a DSC of 84.5% on seven OARs on average, higher than RB or DL alone by 4.5% and 4.7%. The DSC of Def-RgDL is 86.5%, higher than DIR or DL alone by 2.4% and 6.7%. The inference time took by the DL model to generate final segmentations of seven OARs is less than one second in RgDL. The resulting segmentation accuracy and efficiency show the promise of applying RgDL framework for online ART.
Deep Reinforcement Learning in a Monetary Model
Apr 19, 2021
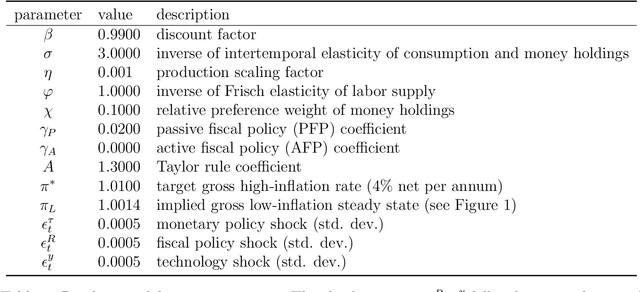


Abstract:We propose using deep reinforcement learning to solve dynamic stochastic general equilibrium models. Agents are represented by deep artificial neural networks and learn to solve their dynamic optimisation problem by interacting with the model environment, of which they have no a priori knowledge. Deep reinforcement learning offers a flexible yet principled way to model bounded rationality within this general class of models. We apply our proposed approach to a classical model from the adaptive learning literature in macroeconomics which looks at the interaction of monetary and fiscal policy. We find that, contrary to adaptive learning, the artificially intelligent household can solve the model in all policy regimes.
Breast Ultrasound Computer-Aided Diagnosis Using Structure-Aware Triplet Path Networks
Aug 09, 2019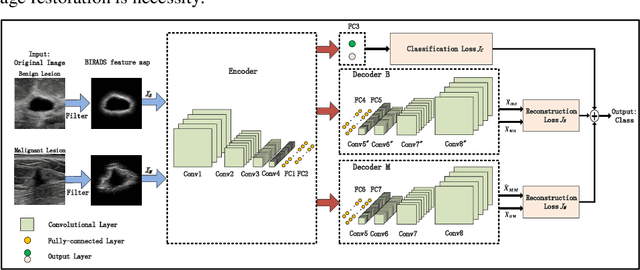
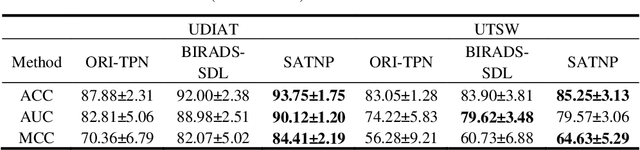
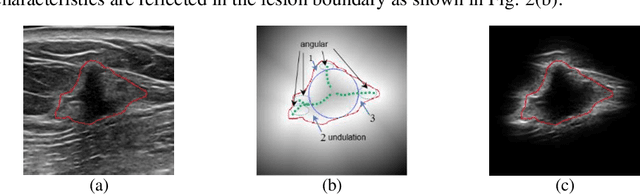
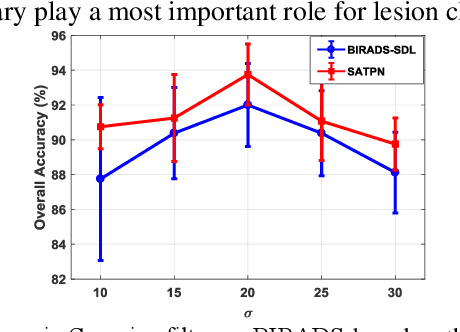
Abstract:Breast ultrasound (US) is an effective imaging modality for breast cancer detec-tion and diagnosis. The structural characteristics of breast lesion play an im-portant role in Computer-Aided Diagnosis (CAD). In this paper, a novel struc-ture-aware triplet path networks (SATPN) was designed to integrate classifica-tion and two image reconstruction tasks to achieve accurate diagnosis on US im-ages with small training dataset. Specifically, we enhance clinically-approved breast lesion structure characteristics though converting original breast US imag-es to BIRADS-oriented feature maps (BFMs) with a distance-transformation coupled Gaussian filter. Then, the converted BFMs were used as the inputs of SATPN, which performed lesion classification task and two unsupervised stacked convolutional Auto-Encoder (SCAE) networks for benign and malignant image reconstruction tasks, independently. We trained the SATPN with an alter-native learning strategy by balancing image reconstruction error and classification label prediction error. At the test stage, the lesion label was determined by the weighted voting with reconstruction error and label prediction error. We com-pared the performance of the SATPN with TPN using original image as input and our previous developed semi-supervised deep learning methods using BFMs as inputs. Experimental results on two breast US datasets showed that SATPN ranked the best among the three networks, with classification accuracy around 93.5%. These findings indicated that SATPN is promising for effective breast US lesion CAD using small datasets.
Towards automated patient data cleaning using deep learning: A feasibility study on the standardization of organ labeling
Dec 30, 2017



Abstract:Data cleaning consumes about 80% of the time spent on data analysis for clinical research projects. This is a much bigger problem in the era of big data and machine learning in the field of medicine where large volumes of data are being generated. We report an initial effort towards automated patient data cleaning using deep learning: the standardization of organ labeling in radiation therapy. Organs are often labeled inconsistently at different institutions (sometimes even within the same institution) and at different time periods, which poses a problem for clinical research, especially for multi-institutional collaborative clinical research where the acquired patient data is not being used effectively. We developed a convolutional neural network (CNN) to automatically identify each organ in the CT image and then label it with the standardized nomenclature presented at AAPM Task Group 263. We tested this model on the CT images of 54 patients with prostate and 100 patients with head and neck cancer who previously received radiation therapy. The model achieved 100% accuracy in detecting organs and assigning standardized labels for the patients tested. This work shows the feasibility of using deep learning in patient data cleaning that enables standardized datasets to be generated for effective intra- and interinstitutional collaborative clinical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge